Pre Gene Modal
BGIBMGA013140
Annotation
short-chain_specific_acyl-CoA_dehydrogenase_[Agrotis_segetum]
Location in the cell
Mitochondrial Reliability : 2.715
Sequence
CDS
ATGTTTATGCCTTTCTCTAAAACCATTCAAGCTTTAGGTCAATCAGTCACTAGACATACAAGATGCATCGCTTCTTTATCAGCATTATCTGAAGATTATCAGATGTTGCAAAAAACATGCCGTGATTTTGCGGAAGGCGAACTTAAGCCCAATGCTGCTAAATACGATCGAGATCATTTATATCCCGGAGAAGCAATTAAAAAATTAGGAGAACTAGGCTTAATGGCTATAGCGGTACCGGAAAAATGGGGAGGTGCAGGCTTAGACTATTTAGCATACGCTATAGCTCTCGAAGAGATATCCAGAGGTTGTGCTTCAGCTGGTGTTATAATGTCAGTGAATAACTCCTTATATCTTGGACCGCTGTCTCATTGGGGCACTGACAAACAAAAAGAACAGTTTGTAACTCCTTTTTGTACTGGTGATCCTGTGGGCTGCTTTGCTCTATCAGAACCCGGTAATGGATCAGATGCAGGAGCCGCCTCCACTACAGCAAAAGATGCGGGTGACAAGTGGATTCTGAATGGAACTAAATGTTGGATTACCAATGGATACGAGAGCAAAGCATCAGTAGTTTTTGCTACAACTGATAAGAGTTTAAAACATAAAGGTATTTCAGCATTTGTAGTTCCAAAACCTATAAAGGGTCTTGAATTAGGGAAAAAAGAAGATAAACTAGGTATAAGGGGGAGTTCAACTTGCTCCCTGATATATGAAGACTGTGAAATTCCCAAAGAGAACATCCTGGGTGAACCGGGGTTTGGATTCAAGATAGCTATGATGACTCTAGATGCGGGGAGAATTGGTATTGCTTCACAAGCTTTAGGTATAGCTCAGGCTTCACTGGATGTGGCAATAGAATATGCTTCGAAACGCATTGCTTTTGGCAAACCAATATTGAAATTGCAAGCTATTCAAAATAAATTAGCTGAAATGGCGCTCCGTCTTGAATCTGCACGCTTGCTTACATGGCGAGCGGCCTGGCTGAAGGACAACAAAAATCCCTATACTAAGGAGGCAGCAATGGCCAAGTTGGCTGCATCAGAGGCTGCTACGTTTTTATCACACCAATGTATCCAGATTCTTGGAGGAATGGGTTATGTATCAGATATGCCAGCAGAACGACATTACCGCGACGCTCGCATCACTGAGATTTACGAGGGCACCAGCGAAATACAAAGACTGGTTATAGCAGGACAACTGATCAAGGAATACGGACTGAATTAG
Protein
MFMPFSKTIQALGQSVTRHTRCIASLSALSEDYQMLQKTCRDFAEGELKPNAAKYDRDHLYPGEAIKKLGELGLMAIAVPEKWGGAGLDYLAYAIALEEISRGCASAGVIMSVNNSLYLGPLSHWGTDKQKEQFVTPFCTGDPVGCFALSEPGNGSDAGAASTTAKDAGDKWILNGTKCWITNGYESKASVVFATTDKSLKHKGISAFVVPKPIKGLELGKKEDKLGIRGSSTCSLIYEDCEIPKENILGEPGFGFKIAMMTLDAGRIGIASQALGIAQASLDVAIEYASKRIAFGKPILKLQAIQNKLAEMALRLESARLLTWRAAWLKDNKNPYTKEAAMAKLAASEAATFLSHQCIQILGGMGYVSDMPAERHYRDARITEIYEGTSEIQRLVIAGQLIKEYGLN
Summary
Cofactor
FAD
Similarity
Belongs to the acyl-CoA dehydrogenase family.
Uniprot
H9JUC7
A0A068FL57
A0A2A4K8V0
A0A2H1V0Y7
A0A3S2LN21
A0A0N1IPD1
+ More
S4P5F9 A0A212F271 A0A194QI36 V5I817 D2A4V5 A0A182XBG8 A0A182I0A8 A0A182MPT9 A0A182VKN3 N6TTA1 A0A182TPS4 Q7PYI1 A0A182JR73 A0A182KLX6 Q17DJ8 A0A182RXK0 A0A182PQV0 A0A182F884 A0A2M4AVD0 A0A1B6FF77 A0A1S4F6S3 A0A182Q2T6 A0A0L0CPT7 W5JE72 A0A182GT73 B0WWP9 A0A182VR31 A0A1I8N4U0 A0A182JK17 A0A1I8P8R5 A0A336MX34 A0A182YG93 A0A2M4BR67 A0A1Y9H2B2 A0A336MN59 B4NGE2 A0A1Q3F8Y5 Q8I184 A0A0A9XD73 A0A3B0K6K4 A0A084VS48 A0A0K8TFV8 B4M4S1 T1PP05 A0A1B0D978 B4IHR3 Q294T1 B4GMN1 A0A0M4F5W0 A0A182S8A9 A0A1W4W979 Q9VDT1 B4QST3 J9K8V9 B3P075 U5ERH0 A0A1A9Z6I1 A0A1J1J5M8 A0A2J7R9D2 A0A2J7R9F2 D3TNL8 B4PNK9 U4TRD2 A0A1A9V0D8 A0A1B6DC32 Q8I157 A0A1A9YI27 A0A1B0C4K6 B4K4S0 A0A182GCL6 T1HJ19 B3LXS8 B4JT09 A0A1L8E2B4 A0A1L8E378 A0A0P4VP28 A0A1Y1M7X6 E9IJI1 A0A0P5GEL1 A0A0P5WLT9 A0A0P6BFB1 A0A1D2NJT3 A0A0P5WRD3 A0A0P6GUT6 A0A164XQW5 A0A1B0GIU4 A0A3L8DUZ4 A0A026W460 E9GLS5 A0A0N7ZAQ3 A0A195FCJ9 F4X889 A0A195DR28 A0A195BF30
S4P5F9 A0A212F271 A0A194QI36 V5I817 D2A4V5 A0A182XBG8 A0A182I0A8 A0A182MPT9 A0A182VKN3 N6TTA1 A0A182TPS4 Q7PYI1 A0A182JR73 A0A182KLX6 Q17DJ8 A0A182RXK0 A0A182PQV0 A0A182F884 A0A2M4AVD0 A0A1B6FF77 A0A1S4F6S3 A0A182Q2T6 A0A0L0CPT7 W5JE72 A0A182GT73 B0WWP9 A0A182VR31 A0A1I8N4U0 A0A182JK17 A0A1I8P8R5 A0A336MX34 A0A182YG93 A0A2M4BR67 A0A1Y9H2B2 A0A336MN59 B4NGE2 A0A1Q3F8Y5 Q8I184 A0A0A9XD73 A0A3B0K6K4 A0A084VS48 A0A0K8TFV8 B4M4S1 T1PP05 A0A1B0D978 B4IHR3 Q294T1 B4GMN1 A0A0M4F5W0 A0A182S8A9 A0A1W4W979 Q9VDT1 B4QST3 J9K8V9 B3P075 U5ERH0 A0A1A9Z6I1 A0A1J1J5M8 A0A2J7R9D2 A0A2J7R9F2 D3TNL8 B4PNK9 U4TRD2 A0A1A9V0D8 A0A1B6DC32 Q8I157 A0A1A9YI27 A0A1B0C4K6 B4K4S0 A0A182GCL6 T1HJ19 B3LXS8 B4JT09 A0A1L8E2B4 A0A1L8E378 A0A0P4VP28 A0A1Y1M7X6 E9IJI1 A0A0P5GEL1 A0A0P5WLT9 A0A0P6BFB1 A0A1D2NJT3 A0A0P5WRD3 A0A0P6GUT6 A0A164XQW5 A0A1B0GIU4 A0A3L8DUZ4 A0A026W460 E9GLS5 A0A0N7ZAQ3 A0A195FCJ9 F4X889 A0A195DR28 A0A195BF30
Pubmed
19121390
26385554
26354079
23622113
22118469
18362917
+ More
19820115 23537049 12364791 14747013 17210077 20966253 17510324 26108605 20920257 23761445 26483478 25315136 25244985 17994087 25401762 26823975 24438588 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20353571 17550304 12537575 27129103 28004739 21282665 27289101 30249741 24508170 21292972 21719571
19820115 23537049 12364791 14747013 17210077 20966253 17510324 26108605 20920257 23761445 26483478 25315136 25244985 17994087 25401762 26823975 24438588 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20353571 17550304 12537575 27129103 28004739 21282665 27289101 30249741 24508170 21292972 21719571
EMBL
BABH01038556
KJ622078
AID66668.1
NWSH01000035
PCG80426.1
ODYU01000178
+ More
SOQ34523.1 RSAL01000051 RVE50229.1 KQ460426 KPJ14794.1 GAIX01005569 JAA86991.1 AGBW02010783 OWR47848.1 KQ458756 KPJ05223.1 GALX01005465 JAB63001.1 KQ971344 EFA05271.2 APCN01002058 AXCM01003378 APGK01055160 KB741261 ENN71561.1 AAAB01008987 EAA01272.5 CH477293 EAT44503.1 GGFK01011351 MBW44672.1 GECZ01020922 GECZ01006541 JAS48847.1 JAS63228.1 AXCN02000031 JRES01000091 KNC34207.1 ADMH02001488 ETN62356.1 JXUM01086406 JXUM01086407 KQ563565 KXJ73655.1 DS232151 EDS36141.1 UFQS01002855 UFQT01002855 SSX14769.1 SSX34161.1 GGFJ01006353 MBW55494.1 UFQS01001439 UFQT01001439 SSX10901.1 SSX30579.1 CH964251 EDW83359.1 GFDL01011015 JAV24030.1 AY190957 AAO01087.1 GBHO01034606 GBHO01025680 GBHO01025675 GBHO01025655 GBHO01025636 GBHO01005625 GDHC01019463 JAG08998.1 JAG17924.1 JAG17929.1 JAG17949.1 JAG17968.1 JAG37979.1 JAP99165.1 OUUW01000008 SPP83700.1 ATLV01015810 KE525036 KFB40792.1 GBRD01001832 GBRD01001831 JAG63990.1 CH940652 EDW59632.1 KA649723 AFP64352.1 AJVK01004271 CH480840 EDW49430.1 CM000070 EAL28883.2 CH479185 EDW38105.1 CP012526 ALC47337.1 AE014297 AY119269 AAF55709.1 AAM51129.1 CM000364 EDX12293.1 ABLF02025276 CH954181 EDV48449.1 GANO01002811 JAB57060.1 CVRI01000070 CRL07274.1 NEVH01006596 PNF37450.1 PNF37453.1 EZ423020 ADD19296.1 CM000160 EDW96072.1 KB631024 KB631053 ERL84019.1 ERL84047.1 GEDC01023714 GEDC01023145 GEDC01014059 GEDC01006095 JAS13584.1 JAS14153.1 JAS23239.1 JAS31203.1 AY190960 AAO01112.1 JXJN01025526 CH933806 EDW16073.1 JXUM01009662 KQ560288 KXJ83246.1 ACPB03006494 CH902617 EDV41735.1 CH916373 EDV94899.1 GFDF01001201 JAV12883.1 GFDF01001139 JAV12945.1 GDKW01001847 JAI54748.1 GEZM01039896 JAV81108.1 GL763802 EFZ19288.1 GDIQ01249301 JAK02424.1 GDIP01084396 JAM19319.1 GDIP01015571 JAM88144.1 LJIJ01000022 ODN05497.1 GDIP01082593 JAM21122.1 GDIQ01028522 JAN66215.1 LRGB01000944 KZS14477.1 AJWK01017657 QOIP01000004 RLU23709.1 KK107459 EZA50391.1 GL732551 EFX79557.1 GDRN01095577 JAI59488.1 KQ981673 KYN38365.1 GL888932 EGI57190.1 KQ980581 KYN15267.1 KQ976500 KYM83186.1
SOQ34523.1 RSAL01000051 RVE50229.1 KQ460426 KPJ14794.1 GAIX01005569 JAA86991.1 AGBW02010783 OWR47848.1 KQ458756 KPJ05223.1 GALX01005465 JAB63001.1 KQ971344 EFA05271.2 APCN01002058 AXCM01003378 APGK01055160 KB741261 ENN71561.1 AAAB01008987 EAA01272.5 CH477293 EAT44503.1 GGFK01011351 MBW44672.1 GECZ01020922 GECZ01006541 JAS48847.1 JAS63228.1 AXCN02000031 JRES01000091 KNC34207.1 ADMH02001488 ETN62356.1 JXUM01086406 JXUM01086407 KQ563565 KXJ73655.1 DS232151 EDS36141.1 UFQS01002855 UFQT01002855 SSX14769.1 SSX34161.1 GGFJ01006353 MBW55494.1 UFQS01001439 UFQT01001439 SSX10901.1 SSX30579.1 CH964251 EDW83359.1 GFDL01011015 JAV24030.1 AY190957 AAO01087.1 GBHO01034606 GBHO01025680 GBHO01025675 GBHO01025655 GBHO01025636 GBHO01005625 GDHC01019463 JAG08998.1 JAG17924.1 JAG17929.1 JAG17949.1 JAG17968.1 JAG37979.1 JAP99165.1 OUUW01000008 SPP83700.1 ATLV01015810 KE525036 KFB40792.1 GBRD01001832 GBRD01001831 JAG63990.1 CH940652 EDW59632.1 KA649723 AFP64352.1 AJVK01004271 CH480840 EDW49430.1 CM000070 EAL28883.2 CH479185 EDW38105.1 CP012526 ALC47337.1 AE014297 AY119269 AAF55709.1 AAM51129.1 CM000364 EDX12293.1 ABLF02025276 CH954181 EDV48449.1 GANO01002811 JAB57060.1 CVRI01000070 CRL07274.1 NEVH01006596 PNF37450.1 PNF37453.1 EZ423020 ADD19296.1 CM000160 EDW96072.1 KB631024 KB631053 ERL84019.1 ERL84047.1 GEDC01023714 GEDC01023145 GEDC01014059 GEDC01006095 JAS13584.1 JAS14153.1 JAS23239.1 JAS31203.1 AY190960 AAO01112.1 JXJN01025526 CH933806 EDW16073.1 JXUM01009662 KQ560288 KXJ83246.1 ACPB03006494 CH902617 EDV41735.1 CH916373 EDV94899.1 GFDF01001201 JAV12883.1 GFDF01001139 JAV12945.1 GDKW01001847 JAI54748.1 GEZM01039896 JAV81108.1 GL763802 EFZ19288.1 GDIQ01249301 JAK02424.1 GDIP01084396 JAM19319.1 GDIP01015571 JAM88144.1 LJIJ01000022 ODN05497.1 GDIP01082593 JAM21122.1 GDIQ01028522 JAN66215.1 LRGB01000944 KZS14477.1 AJWK01017657 QOIP01000004 RLU23709.1 KK107459 EZA50391.1 GL732551 EFX79557.1 GDRN01095577 JAI59488.1 KQ981673 KYN38365.1 GL888932 EGI57190.1 KQ980581 KYN15267.1 KQ976500 KYM83186.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000007151
UP000053268
+ More
UP000007266 UP000076407 UP000075840 UP000075883 UP000075903 UP000019118 UP000075902 UP000007062 UP000075881 UP000075882 UP000008820 UP000075900 UP000075885 UP000069272 UP000075886 UP000037069 UP000000673 UP000069940 UP000249989 UP000002320 UP000075920 UP000095301 UP000075880 UP000095300 UP000076408 UP000075884 UP000007798 UP000268350 UP000030765 UP000008792 UP000092462 UP000001292 UP000001819 UP000008744 UP000092553 UP000075901 UP000192221 UP000000803 UP000000304 UP000007819 UP000008711 UP000092445 UP000183832 UP000235965 UP000002282 UP000030742 UP000078200 UP000092443 UP000092460 UP000009192 UP000015103 UP000007801 UP000001070 UP000094527 UP000076858 UP000092461 UP000279307 UP000053097 UP000000305 UP000078541 UP000007755 UP000078492 UP000078540
UP000007266 UP000076407 UP000075840 UP000075883 UP000075903 UP000019118 UP000075902 UP000007062 UP000075881 UP000075882 UP000008820 UP000075900 UP000075885 UP000069272 UP000075886 UP000037069 UP000000673 UP000069940 UP000249989 UP000002320 UP000075920 UP000095301 UP000075880 UP000095300 UP000076408 UP000075884 UP000007798 UP000268350 UP000030765 UP000008792 UP000092462 UP000001292 UP000001819 UP000008744 UP000092553 UP000075901 UP000192221 UP000000803 UP000000304 UP000007819 UP000008711 UP000092445 UP000183832 UP000235965 UP000002282 UP000030742 UP000078200 UP000092443 UP000092460 UP000009192 UP000015103 UP000007801 UP000001070 UP000094527 UP000076858 UP000092461 UP000279307 UP000053097 UP000000305 UP000078541 UP000007755 UP000078492 UP000078540
Pfam
Interpro
IPR006091
Acyl-CoA_Oxase/DH_cen-dom
+ More
IPR009100 AcylCoA_DH/oxidase_NM_dom
IPR009075 AcylCo_DH/oxidase_C
IPR006089 Acyl-CoA_DH_CS
IPR013786 AcylCoA_DH/ox_N
IPR037069 AcylCoA_DH/ox_N_sf
IPR036250 AcylCo_DH-like_C
IPR013107 Acyl-CoA_DH_C_dom
IPR000819 Peptidase_M17_C
IPR008283 Peptidase_M17_N
IPR011356 Leucine_aapep/pepB
IPR009100 AcylCoA_DH/oxidase_NM_dom
IPR009075 AcylCo_DH/oxidase_C
IPR006089 Acyl-CoA_DH_CS
IPR013786 AcylCoA_DH/ox_N
IPR037069 AcylCoA_DH/ox_N_sf
IPR036250 AcylCo_DH-like_C
IPR013107 Acyl-CoA_DH_C_dom
IPR000819 Peptidase_M17_C
IPR008283 Peptidase_M17_N
IPR011356 Leucine_aapep/pepB
Gene 3D
ProteinModelPortal
H9JUC7
A0A068FL57
A0A2A4K8V0
A0A2H1V0Y7
A0A3S2LN21
A0A0N1IPD1
+ More
S4P5F9 A0A212F271 A0A194QI36 V5I817 D2A4V5 A0A182XBG8 A0A182I0A8 A0A182MPT9 A0A182VKN3 N6TTA1 A0A182TPS4 Q7PYI1 A0A182JR73 A0A182KLX6 Q17DJ8 A0A182RXK0 A0A182PQV0 A0A182F884 A0A2M4AVD0 A0A1B6FF77 A0A1S4F6S3 A0A182Q2T6 A0A0L0CPT7 W5JE72 A0A182GT73 B0WWP9 A0A182VR31 A0A1I8N4U0 A0A182JK17 A0A1I8P8R5 A0A336MX34 A0A182YG93 A0A2M4BR67 A0A1Y9H2B2 A0A336MN59 B4NGE2 A0A1Q3F8Y5 Q8I184 A0A0A9XD73 A0A3B0K6K4 A0A084VS48 A0A0K8TFV8 B4M4S1 T1PP05 A0A1B0D978 B4IHR3 Q294T1 B4GMN1 A0A0M4F5W0 A0A182S8A9 A0A1W4W979 Q9VDT1 B4QST3 J9K8V9 B3P075 U5ERH0 A0A1A9Z6I1 A0A1J1J5M8 A0A2J7R9D2 A0A2J7R9F2 D3TNL8 B4PNK9 U4TRD2 A0A1A9V0D8 A0A1B6DC32 Q8I157 A0A1A9YI27 A0A1B0C4K6 B4K4S0 A0A182GCL6 T1HJ19 B3LXS8 B4JT09 A0A1L8E2B4 A0A1L8E378 A0A0P4VP28 A0A1Y1M7X6 E9IJI1 A0A0P5GEL1 A0A0P5WLT9 A0A0P6BFB1 A0A1D2NJT3 A0A0P5WRD3 A0A0P6GUT6 A0A164XQW5 A0A1B0GIU4 A0A3L8DUZ4 A0A026W460 E9GLS5 A0A0N7ZAQ3 A0A195FCJ9 F4X889 A0A195DR28 A0A195BF30
S4P5F9 A0A212F271 A0A194QI36 V5I817 D2A4V5 A0A182XBG8 A0A182I0A8 A0A182MPT9 A0A182VKN3 N6TTA1 A0A182TPS4 Q7PYI1 A0A182JR73 A0A182KLX6 Q17DJ8 A0A182RXK0 A0A182PQV0 A0A182F884 A0A2M4AVD0 A0A1B6FF77 A0A1S4F6S3 A0A182Q2T6 A0A0L0CPT7 W5JE72 A0A182GT73 B0WWP9 A0A182VR31 A0A1I8N4U0 A0A182JK17 A0A1I8P8R5 A0A336MX34 A0A182YG93 A0A2M4BR67 A0A1Y9H2B2 A0A336MN59 B4NGE2 A0A1Q3F8Y5 Q8I184 A0A0A9XD73 A0A3B0K6K4 A0A084VS48 A0A0K8TFV8 B4M4S1 T1PP05 A0A1B0D978 B4IHR3 Q294T1 B4GMN1 A0A0M4F5W0 A0A182S8A9 A0A1W4W979 Q9VDT1 B4QST3 J9K8V9 B3P075 U5ERH0 A0A1A9Z6I1 A0A1J1J5M8 A0A2J7R9D2 A0A2J7R9F2 D3TNL8 B4PNK9 U4TRD2 A0A1A9V0D8 A0A1B6DC32 Q8I157 A0A1A9YI27 A0A1B0C4K6 B4K4S0 A0A182GCL6 T1HJ19 B3LXS8 B4JT09 A0A1L8E2B4 A0A1L8E378 A0A0P4VP28 A0A1Y1M7X6 E9IJI1 A0A0P5GEL1 A0A0P5WLT9 A0A0P6BFB1 A0A1D2NJT3 A0A0P5WRD3 A0A0P6GUT6 A0A164XQW5 A0A1B0GIU4 A0A3L8DUZ4 A0A026W460 E9GLS5 A0A0N7ZAQ3 A0A195FCJ9 F4X889 A0A195DR28 A0A195BF30
PDB
1JQI
E-value=8.47208e-150,
Score=1360
Ontologies
PATHWAY
00071
Fatty acid degradation - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00640 Propanoate metabolism - Bombyx mori (domestic silkworm)
00650 Butanoate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01212 Fatty acid metabolism - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00640 Propanoate metabolism - Bombyx mori (domestic silkworm)
00650 Butanoate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01212 Fatty acid metabolism - Bombyx mori (domestic silkworm)
GO
PANTHER
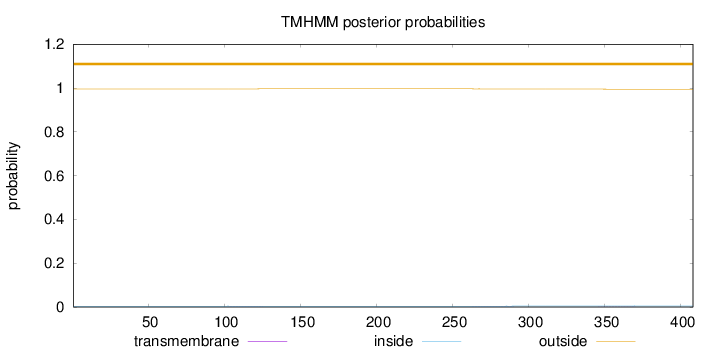
Topology
Length:
408
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04216
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00387
outside
1 - 408
Population Genetic Test Statistics
Pi
213.795328
Theta
177.542506
Tajima's D
0.577513
CLR
0.122552
CSRT
0.539023048847558
Interpretation
Uncertain
