Gene
KWMTBOMO09813
Annotation
PREDICTED:_uncharacterized_protein_LOC101747036_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.28
Sequence
CDS
ATGCCAAATGGCAACATATTCAGAGGAAAAGAACGTCTAGTAAAACAAGTTGAACCAAAACATTTACGGCGTATTAAAGAAGATTTTGCCATTGAGGAACAAAATATGCTTTATTTACGTTTCCCCTATTTGTCCGAGGCAGAAAGTTTTGGACACACAAAAGCTTTAGGAAAACATGAAATGAGAAAAGAAATGCTCAATGACAAGAATAGAAGAATATTTAAACAAGATGTAACATTATATGAACGATTACAGCACCTAAGAATCGGCGAAAAATGGGAGTAA
Protein
MPNGNIFRGKERLVKQVEPKHLRRIKEDFAIEEQNMLYLRFPYLSEAESFGHTKALGKHEMRKEMLNDKNRRIFKQDVTLYERLQHLRIGEKWE
Summary
Uniprot
A0A2A4K9T1
A0A2H1V111
A0A194QIK5
A0A0L7KUV4
A0A212FJ48
S4PS24
+ More
A0A0N0PD81 A0A1E1VXF7 A0A437BIP5 A0A1W4XED0 W8BL33 A0A0K8VBF9 A0A034W2Q9 A0A1L8DSF7 A0A0C9S0Z9 N6UPJ9 R4WNN4 A0A0L0CKB0 V5GPQ6 A0A1I8MJ04 K7ITA6 A0A1B0D772 A0A336LRH6 A0A336L4Q7 A0A1L8EDR6 A0A1B0CA55 A0A1I8PUD3 A0A1A9VWW9 A0A2R7W1T2 A0A1A9Z3Y0 A0A026WAM4 A0A2P8XGA3 A0A232FCZ2 A0A067RKA4 A0A1B6GNS3 A0A195FC60 A0A1A9XM12 A0A1B0BAU4 A0A1D2MRI0 B4L5W4 F4WL46 A0A1Y1LLD9 A0A1Q3F9F2 A0A158NZK2 Q9W551 O97419 A0A182MKT9 A0A1B6MRS8 A0A1D2NM69 A0A182RBW9 A0A182N6V3 A0A182PBE9 W5JJ75 A0A151WLU3 B4M7F5 B4R315 B4I9J8 A0A182VQD8 A0A195CJ26 A0A195B625 A0A2C9H3S4 A0A182V2S6 A0A182TMJ0 A0A182WYH6 A0A182I8H8 A0A2M4C496 A0A2M4AQS6 B4JMX7 B4N237 A0A0P5FAX4 D6WYS1 B0X5M2 A0A1W4VRK0 U5EU22 A0A0P4WT07 A0A1J1IFP0 A0A182K0W4 A0A2M3ZB17 A0A182Q1B1 B4H343 Q16QW0 A0A084VBU7 B3P9N2 B4Q1L3 A0A1B6I8D5 A0A3B0J976 B4PY71 A0A182FNP9 E9G6U3 B3MYY9 A0A1A9WA20 A0A195EK93 A0A182XX99 Q29JJ8 D0QWG3 J9L3V1 A0A182SCD7 A0A2S2P6D9 A0A182YTC4
A0A0N0PD81 A0A1E1VXF7 A0A437BIP5 A0A1W4XED0 W8BL33 A0A0K8VBF9 A0A034W2Q9 A0A1L8DSF7 A0A0C9S0Z9 N6UPJ9 R4WNN4 A0A0L0CKB0 V5GPQ6 A0A1I8MJ04 K7ITA6 A0A1B0D772 A0A336LRH6 A0A336L4Q7 A0A1L8EDR6 A0A1B0CA55 A0A1I8PUD3 A0A1A9VWW9 A0A2R7W1T2 A0A1A9Z3Y0 A0A026WAM4 A0A2P8XGA3 A0A232FCZ2 A0A067RKA4 A0A1B6GNS3 A0A195FC60 A0A1A9XM12 A0A1B0BAU4 A0A1D2MRI0 B4L5W4 F4WL46 A0A1Y1LLD9 A0A1Q3F9F2 A0A158NZK2 Q9W551 O97419 A0A182MKT9 A0A1B6MRS8 A0A1D2NM69 A0A182RBW9 A0A182N6V3 A0A182PBE9 W5JJ75 A0A151WLU3 B4M7F5 B4R315 B4I9J8 A0A182VQD8 A0A195CJ26 A0A195B625 A0A2C9H3S4 A0A182V2S6 A0A182TMJ0 A0A182WYH6 A0A182I8H8 A0A2M4C496 A0A2M4AQS6 B4JMX7 B4N237 A0A0P5FAX4 D6WYS1 B0X5M2 A0A1W4VRK0 U5EU22 A0A0P4WT07 A0A1J1IFP0 A0A182K0W4 A0A2M3ZB17 A0A182Q1B1 B4H343 Q16QW0 A0A084VBU7 B3P9N2 B4Q1L3 A0A1B6I8D5 A0A3B0J976 B4PY71 A0A182FNP9 E9G6U3 B3MYY9 A0A1A9WA20 A0A195EK93 A0A182XX99 Q29JJ8 D0QWG3 J9L3V1 A0A182SCD7 A0A2S2P6D9 A0A182YTC4
Pubmed
26354079
26227816
22118469
23622113
24495485
25348373
+ More
23537049 23691247 26108605 25315136 20075255 24508170 30249741 29403074 28648823 24845553 27289101 17994087 21719571 28004739 21347285 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 12364791 18362917 19820115 17510324 24438588 17550304 21292972 25244985 15632085 19859648
23537049 23691247 26108605 25315136 20075255 24508170 30249741 29403074 28648823 24845553 27289101 17994087 21719571 28004739 21347285 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 12364791 18362917 19820115 17510324 24438588 17550304 21292972 25244985 15632085 19859648
EMBL
NWSH01000035
PCG80422.1
ODYU01000178
SOQ34527.1
KQ458756
KPJ05229.1
+ More
JTDY01005590 KOB66851.1 AGBW02008320 OWR53755.1 GAIX01014233 JAA78327.1 KQ460426 KPJ14798.1 GDQN01011690 JAT79364.1 RSAL01000051 RVE50233.1 GAMC01016216 JAB90339.1 GDHF01016030 JAI36284.1 GAKP01009968 GAKP01009965 GAKP01009964 JAC48984.1 GFDF01004767 JAV09317.1 GBYB01014336 JAG84103.1 APGK01022718 KB740442 KB632185 ENN80647.1 ERL89664.1 AK417266 BAN20481.1 JRES01000290 KNC32701.1 GALX01002362 JAB66104.1 AAZX01008304 AJVK01012351 UFQT01000056 SSX19129.1 UFQS01001992 UFQT01001992 SSX12896.1 SSX32338.1 GFDG01002050 JAV16749.1 AJWK01003289 KK854247 PTY13683.1 KK107353 QOIP01000012 EZA52079.1 RLU15756.1 PYGN01002250 PSN31005.1 NNAY01000418 OXU28545.1 KK852575 KDR21036.1 GECZ01005702 JAS64067.1 KQ981685 KYN37998.1 JXJN01011153 LJIJ01000634 ODM95687.1 CH933811 EDW06573.1 GL888207 EGI65009.1 GEZM01052510 JAV74469.1 GFDL01010855 JAV24190.1 ADTU01004711 AE014298 BT126349 KX531521 AAF45680.1 AEC11521.1 AHN59267.1 ANY27331.1 AL021106 CAA15941.1 AXCM01006229 GEBQ01025488 GEBQ01001335 GEBQ01000508 JAT14489.1 JAT38642.1 JAT39469.1 LJIJ01000009 ODN06185.1 ADMH02001019 ETN64402.1 KQ982944 KYQ48862.1 CH940653 EDW62722.1 CM000366 EDX16869.1 CH480825 EDW43879.1 KQ977720 KYN00437.1 KQ976587 KYM79710.1 AAAB01008805 APCN01003033 GGFJ01011006 MBW60147.1 GGFK01009813 MBW43134.1 CH916371 EDV92070.1 CH963925 EDW78426.1 GDIQ01257161 LRGB01001579 JAJ94563.1 KZS11538.1 KQ971357 EFA08456.1 DS232386 EDS40959.1 GANO01002525 JAB57346.1 GDRN01025713 JAI67708.1 CVRI01000047 CRK97830.1 GGFM01004983 MBW25734.1 AXCN02000309 CH479205 EDW30736.1 CH477729 EAT36791.1 ATLV01009527 KE524533 KFB35441.1 CH954183 EDV45528.1 CM000162 EDX01454.1 GECU01024505 JAS83201.1 OUUW01000003 SPP78814.1 EDX00944.1 GL732533 EFX84804.1 CH902632 EDV32833.1 KQ978747 KYN28680.1 CH379063 EAL32303.1 FJ821027 ACN94655.1 ABLF02025591 GGMR01011847 MBY24466.1
JTDY01005590 KOB66851.1 AGBW02008320 OWR53755.1 GAIX01014233 JAA78327.1 KQ460426 KPJ14798.1 GDQN01011690 JAT79364.1 RSAL01000051 RVE50233.1 GAMC01016216 JAB90339.1 GDHF01016030 JAI36284.1 GAKP01009968 GAKP01009965 GAKP01009964 JAC48984.1 GFDF01004767 JAV09317.1 GBYB01014336 JAG84103.1 APGK01022718 KB740442 KB632185 ENN80647.1 ERL89664.1 AK417266 BAN20481.1 JRES01000290 KNC32701.1 GALX01002362 JAB66104.1 AAZX01008304 AJVK01012351 UFQT01000056 SSX19129.1 UFQS01001992 UFQT01001992 SSX12896.1 SSX32338.1 GFDG01002050 JAV16749.1 AJWK01003289 KK854247 PTY13683.1 KK107353 QOIP01000012 EZA52079.1 RLU15756.1 PYGN01002250 PSN31005.1 NNAY01000418 OXU28545.1 KK852575 KDR21036.1 GECZ01005702 JAS64067.1 KQ981685 KYN37998.1 JXJN01011153 LJIJ01000634 ODM95687.1 CH933811 EDW06573.1 GL888207 EGI65009.1 GEZM01052510 JAV74469.1 GFDL01010855 JAV24190.1 ADTU01004711 AE014298 BT126349 KX531521 AAF45680.1 AEC11521.1 AHN59267.1 ANY27331.1 AL021106 CAA15941.1 AXCM01006229 GEBQ01025488 GEBQ01001335 GEBQ01000508 JAT14489.1 JAT38642.1 JAT39469.1 LJIJ01000009 ODN06185.1 ADMH02001019 ETN64402.1 KQ982944 KYQ48862.1 CH940653 EDW62722.1 CM000366 EDX16869.1 CH480825 EDW43879.1 KQ977720 KYN00437.1 KQ976587 KYM79710.1 AAAB01008805 APCN01003033 GGFJ01011006 MBW60147.1 GGFK01009813 MBW43134.1 CH916371 EDV92070.1 CH963925 EDW78426.1 GDIQ01257161 LRGB01001579 JAJ94563.1 KZS11538.1 KQ971357 EFA08456.1 DS232386 EDS40959.1 GANO01002525 JAB57346.1 GDRN01025713 JAI67708.1 CVRI01000047 CRK97830.1 GGFM01004983 MBW25734.1 AXCN02000309 CH479205 EDW30736.1 CH477729 EAT36791.1 ATLV01009527 KE524533 KFB35441.1 CH954183 EDV45528.1 CM000162 EDX01454.1 GECU01024505 JAS83201.1 OUUW01000003 SPP78814.1 EDX00944.1 GL732533 EFX84804.1 CH902632 EDV32833.1 KQ978747 KYN28680.1 CH379063 EAL32303.1 FJ821027 ACN94655.1 ABLF02025591 GGMR01011847 MBY24466.1
Proteomes
UP000218220
UP000053268
UP000037510
UP000007151
UP000053240
UP000283053
+ More
UP000192223 UP000019118 UP000030742 UP000037069 UP000095301 UP000002358 UP000092462 UP000092461 UP000095300 UP000078200 UP000092445 UP000053097 UP000279307 UP000245037 UP000215335 UP000027135 UP000078541 UP000092443 UP000092460 UP000094527 UP000009192 UP000007755 UP000005205 UP000000803 UP000075883 UP000075900 UP000075884 UP000075885 UP000000673 UP000075809 UP000008792 UP000000304 UP000001292 UP000075920 UP000078542 UP000078540 UP000075903 UP000075902 UP000076407 UP000075840 UP000001070 UP000007798 UP000076858 UP000007266 UP000002320 UP000192221 UP000183832 UP000075881 UP000075886 UP000008744 UP000008820 UP000030765 UP000008711 UP000002282 UP000268350 UP000069272 UP000000305 UP000007801 UP000091820 UP000078492 UP000076408 UP000001819 UP000007819 UP000075901
UP000192223 UP000019118 UP000030742 UP000037069 UP000095301 UP000002358 UP000092462 UP000092461 UP000095300 UP000078200 UP000092445 UP000053097 UP000279307 UP000245037 UP000215335 UP000027135 UP000078541 UP000092443 UP000092460 UP000094527 UP000009192 UP000007755 UP000005205 UP000000803 UP000075883 UP000075900 UP000075884 UP000075885 UP000000673 UP000075809 UP000008792 UP000000304 UP000001292 UP000075920 UP000078542 UP000078540 UP000075903 UP000075902 UP000076407 UP000075840 UP000001070 UP000007798 UP000076858 UP000007266 UP000002320 UP000192221 UP000183832 UP000075881 UP000075886 UP000008744 UP000008820 UP000030765 UP000008711 UP000002282 UP000268350 UP000069272 UP000000305 UP000007801 UP000091820 UP000078492 UP000076408 UP000001819 UP000007819 UP000075901
Pfam
Interpro
IPR016576
Ribosomal_MRP63_mit
+ More
IPR001650 Helicase_C
IPR017884 SANT_dom
IPR009057 Homeobox-like_sf
IPR015194 ISWI_HAND-dom
IPR001005 SANT/Myb
IPR014001 Helicase_ATP-bd
IPR036306 ISWI_HAND-dom_sf
IPR000330 SNF2_N
IPR029915 ISWI_metazoa
IPR015195 SLIDE
IPR027417 P-loop_NTPase
IPR038718 SNF2-like_sf
IPR005958 TyrNic_aminoTrfase
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR004838 NHTrfase_class1_PyrdxlP-BS
IPR015421 PyrdxlP-dep_Trfase_major
IPR004839 Aminotransferase_I/II
IPR005957 Tyrosine_aminoTrfase
IPR015424 PyrdxlP-dep_Trfase
IPR006689 Small_GTPase_ARF/SAR
IPR003734 DUF155
IPR001650 Helicase_C
IPR017884 SANT_dom
IPR009057 Homeobox-like_sf
IPR015194 ISWI_HAND-dom
IPR001005 SANT/Myb
IPR014001 Helicase_ATP-bd
IPR036306 ISWI_HAND-dom_sf
IPR000330 SNF2_N
IPR029915 ISWI_metazoa
IPR015195 SLIDE
IPR027417 P-loop_NTPase
IPR038718 SNF2-like_sf
IPR005958 TyrNic_aminoTrfase
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR004838 NHTrfase_class1_PyrdxlP-BS
IPR015421 PyrdxlP-dep_Trfase_major
IPR004839 Aminotransferase_I/II
IPR005957 Tyrosine_aminoTrfase
IPR015424 PyrdxlP-dep_Trfase
IPR006689 Small_GTPase_ARF/SAR
IPR003734 DUF155
ProteinModelPortal
A0A2A4K9T1
A0A2H1V111
A0A194QIK5
A0A0L7KUV4
A0A212FJ48
S4PS24
+ More
A0A0N0PD81 A0A1E1VXF7 A0A437BIP5 A0A1W4XED0 W8BL33 A0A0K8VBF9 A0A034W2Q9 A0A1L8DSF7 A0A0C9S0Z9 N6UPJ9 R4WNN4 A0A0L0CKB0 V5GPQ6 A0A1I8MJ04 K7ITA6 A0A1B0D772 A0A336LRH6 A0A336L4Q7 A0A1L8EDR6 A0A1B0CA55 A0A1I8PUD3 A0A1A9VWW9 A0A2R7W1T2 A0A1A9Z3Y0 A0A026WAM4 A0A2P8XGA3 A0A232FCZ2 A0A067RKA4 A0A1B6GNS3 A0A195FC60 A0A1A9XM12 A0A1B0BAU4 A0A1D2MRI0 B4L5W4 F4WL46 A0A1Y1LLD9 A0A1Q3F9F2 A0A158NZK2 Q9W551 O97419 A0A182MKT9 A0A1B6MRS8 A0A1D2NM69 A0A182RBW9 A0A182N6V3 A0A182PBE9 W5JJ75 A0A151WLU3 B4M7F5 B4R315 B4I9J8 A0A182VQD8 A0A195CJ26 A0A195B625 A0A2C9H3S4 A0A182V2S6 A0A182TMJ0 A0A182WYH6 A0A182I8H8 A0A2M4C496 A0A2M4AQS6 B4JMX7 B4N237 A0A0P5FAX4 D6WYS1 B0X5M2 A0A1W4VRK0 U5EU22 A0A0P4WT07 A0A1J1IFP0 A0A182K0W4 A0A2M3ZB17 A0A182Q1B1 B4H343 Q16QW0 A0A084VBU7 B3P9N2 B4Q1L3 A0A1B6I8D5 A0A3B0J976 B4PY71 A0A182FNP9 E9G6U3 B3MYY9 A0A1A9WA20 A0A195EK93 A0A182XX99 Q29JJ8 D0QWG3 J9L3V1 A0A182SCD7 A0A2S2P6D9 A0A182YTC4
A0A0N0PD81 A0A1E1VXF7 A0A437BIP5 A0A1W4XED0 W8BL33 A0A0K8VBF9 A0A034W2Q9 A0A1L8DSF7 A0A0C9S0Z9 N6UPJ9 R4WNN4 A0A0L0CKB0 V5GPQ6 A0A1I8MJ04 K7ITA6 A0A1B0D772 A0A336LRH6 A0A336L4Q7 A0A1L8EDR6 A0A1B0CA55 A0A1I8PUD3 A0A1A9VWW9 A0A2R7W1T2 A0A1A9Z3Y0 A0A026WAM4 A0A2P8XGA3 A0A232FCZ2 A0A067RKA4 A0A1B6GNS3 A0A195FC60 A0A1A9XM12 A0A1B0BAU4 A0A1D2MRI0 B4L5W4 F4WL46 A0A1Y1LLD9 A0A1Q3F9F2 A0A158NZK2 Q9W551 O97419 A0A182MKT9 A0A1B6MRS8 A0A1D2NM69 A0A182RBW9 A0A182N6V3 A0A182PBE9 W5JJ75 A0A151WLU3 B4M7F5 B4R315 B4I9J8 A0A182VQD8 A0A195CJ26 A0A195B625 A0A2C9H3S4 A0A182V2S6 A0A182TMJ0 A0A182WYH6 A0A182I8H8 A0A2M4C496 A0A2M4AQS6 B4JMX7 B4N237 A0A0P5FAX4 D6WYS1 B0X5M2 A0A1W4VRK0 U5EU22 A0A0P4WT07 A0A1J1IFP0 A0A182K0W4 A0A2M3ZB17 A0A182Q1B1 B4H343 Q16QW0 A0A084VBU7 B3P9N2 B4Q1L3 A0A1B6I8D5 A0A3B0J976 B4PY71 A0A182FNP9 E9G6U3 B3MYY9 A0A1A9WA20 A0A195EK93 A0A182XX99 Q29JJ8 D0QWG3 J9L3V1 A0A182SCD7 A0A2S2P6D9 A0A182YTC4
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
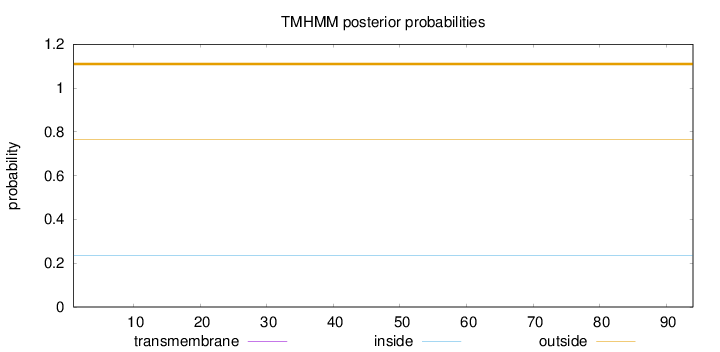
Length:
94
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00018
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.23467
outside
1 - 94
Population Genetic Test Statistics
Pi
17.206817
Theta
16.072812
Tajima's D
-0.053653
CLR
0
CSRT
0.361781910904455
Interpretation
Uncertain
