Gene
KWMTBOMO09812 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013118
Annotation
PREDICTED:_apoptosis-inducing_factor_3_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.711
Sequence
CDS
ATGGGAAGCTCCTACTCGAGATCATATTCTCCCAACAGTGGCCAAGGTGGAAGCTGCAATGAAAACCGCGGAACCATATCCCGAATGGAGTCTACTAACAATTACGTCGAATCAGTCGTTTGTCAAGAGAATGACCTCAAAGACAACGAGATGAAGGTTTTCGACATCGGTGAAGACGGGAAGGTGCTTGTAGTGAAACAGAAGGGAGAATTCTCCGCTATCGGAACGAAGTGTACGCATTATGGAGCCCCTCTCATCAACGGCGCGCTGGGGGACGGAAGACTGAGGTGCCCTTGGCACGGGGCCTGCTTCAATCTGAAGACCGGGGACATTGAAGATTTTCCTGGTTTTGATTCGCTGCCGTGCTATCAAGTTACCGTAACCGATAAAGGAGAAGTAAAGGTAAGAGCAAAGATAAGTGACCTCAAAACGAACAAGCGTATAAAGGATATGGGTGTGGTCTCTCCTTGTGAAGGTTCCGTTGTTGTTATAGTGGGCGGAGGACCGTCGGGCGCTACATGTGCTGAATCACTACGCAGCGAAGGATTTAAAGGTCGAATCACCGTTATCGCAAAAGAGCCGCATCTCCCTTACGATAGGATTAAAGTATCTAAAATCGGCACAGTCACCGATATAGAAAAACTACAGGCGAGATCACAAAAATATTATGACGACGCCAATATTGAAATAATGAAAGGAGTGGAAGCAACCAAAATAGAACCTAACGACAAGCTTGTGCATTTGAACAACGGTAACAAGCTCCGATACAGCGCGCTGTATCTAGCGACCGGCTCCAAGCCTCGGGTACCTGATATACCCGGCATCGATTTGAAAAATATATTTACCGTGCGGAATTATGAAGACTCCATTAACATACTGACAACATTAGGATCGAATCAAGACAAAGACGTTGTCGTCCTCGGTCTCCGTTTCATCGGCTTGGAGTCGGCCGCTTCATGCAATGAGAAAGCGAAGTCCATAACCGTAGTTGGTAAACAGAAGGCTCCGCTTGAGCAAATTTTCGGCCCAGAAATAGGACGAAGCCTACAAAAATTGTTCGAAGATAAAGGCGTAAAATTCCAGTTCGAGACCACGATAACAAAATGTAATGGCGAGGACGGTGTGATAAAGTCGGTCGAATTGACGAACGGGACCGTGCTGCCCGCCGATATGCTAATTTTGGGCGTCGGGACCACATTCAACACAGAATTCGTGAAAGATAGCGGTTTGAAACTGGAAGCGAACGGTGCGGTCTCGGTTAATGATAAGCTCGAAACAAATATTCAAAATATTTACGCCGGAGGTGACATCGCCCATGCACCCGTTGCGGCCTACGATAATAAACAGATGACTATAGGCCACGTGGGATTGTCTCAGTACCACGGACGGGTCGCCGCTCTGAATATATTGAAGAAAGAGACTTCGCTGCAAACTGTACCATTCTTCTGGACAATGTTGTTCGGGAAGTCCATCCGGTACGCCGGCTGCGGTAGGGCCGCAAGCACCCTGATCGATGGAAACGTTGATGATTTAAAATTCGTCATCTTTTTCTTTGACGAAACAGACAAGGTAATCGCCATGGCTTCGTGCATGAGAGATCCCATTGTCGCGCAATTCGCCGAATTCCTACATCAAGGCAAGTCGCTGTACAAGAAAGACTTGAAAGATGATCGATTTGCTTGGACAAAGTCTATCAATTGA
Protein
MGSSYSRSYSPNSGQGGSCNENRGTISRMESTNNYVESVVCQENDLKDNEMKVFDIGEDGKVLVVKQKGEFSAIGTKCTHYGAPLINGALGDGRLRCPWHGACFNLKTGDIEDFPGFDSLPCYQVTVTDKGEVKVRAKISDLKTNKRIKDMGVVSPCEGSVVVIVGGGPSGATCAESLRSEGFKGRITVIAKEPHLPYDRIKVSKIGTVTDIEKLQARSQKYYDDANIEIMKGVEATKIEPNDKLVHLNNGNKLRYSALYLATGSKPRVPDIPGIDLKNIFTVRNYEDSINILTTLGSNQDKDVVVLGLRFIGLESAASCNEKAKSITVVGKQKAPLEQIFGPEIGRSLQKLFEDKGVKFQFETTITKCNGEDGVIKSVELTNGTVLPADMLILGVGTTFNTEFVKDSGLKLEANGAVSVNDKLETNIQNIYAGGDIAHAPVAAYDNKQMTIGHVGLSQYHGRVAALNILKKETSLQTVPFFWTMLFGKSIRYAGCGRAASTLIDGNVDDLKFVIFFFDETDKVIAMASCMRDPIVAQFAEFLHQGKSLYKKDLKDDRFAWTKSIN
Summary
Uniprot
H9JUA5
A0A2H1V112
A0A0L7L1H8
A0A2A4K867
A0A194QJZ1
A0A3S2NG59
+ More
A0A0N1IHV4 A0A212FJ44 A0A336MTC2 A0A336K2W1 A0A182QYX1 A0A084VBU8 A0A182JMA6 A0A182FNP8 A0A1B0D964 A0A182PBE8 A0A182N6V4 W5JJX4 A0A182MSA7 A0A2M4AJ10 A0A2M4AJ71 A0A182K0W3 A0A2M4AJE8 A0A182RBW8 A0A182I8H9 F5HIX4 F7IUV9 A0A182V9F1 U5ESJ0 A0A182WYH5 A0A1J1IGH9 A0A182LAD7 A0A182U109 A0A182VQD7 A0A1W4XEC0 A0A182XX98 A0A1W4XPF0 A0A1Y1NIS5 K7IP01 A0A1L8DJJ8 A0A1Q3F988 B0X5M3 A0A1S4FSQ1 A0A1S4FSV9 A0A310SK92 A0A232EVQ9 Q16QW1 J9HZJ7 E2A429 A0A026W7V4 A0A3L8D5I7 A0A2J7RH83 A0A023EWI7 A0A182G3Z5 A0A2J7RH63 U4UB39 A0A0L7RHE3 E2BWL5 A0A195ELC2 E9IKW0 K7ITA7 A0A0A9Y1F1 A0A146M613 A0A146LSJ3 A0A0A9WCY1 A0A146M5Z0 F4WL47 A0A0A9XGT7 D6WYS2 A0A232FDJ7 A0A1W4XPF5 A0A1W4XE01 A0A1W4XNL8 A0A3B0J912 B4NBZ8 A0A3B0JZF0 A0A3B0JDM7 B4L5W0 D0QWG5 A0A1A9WA16 A0A0R3P265 B4H345 A0A0R3NXK1 A0A0R3NXM4 A0A0R3NY26 B3MYY5 A0A195CIA3 A0A0Q9XF58 A0A0P8XH64 A0A0P9A6W1 A0A224XMI6 A0A1I8MEZ3 A0A151WM05 A0A0R3P2I7 Q29JK0 A0A1I8MEZ5 A0A1A9Z3Y6 O77266 A0A1I8PMJ8 A0A1B6CZA7
A0A0N1IHV4 A0A212FJ44 A0A336MTC2 A0A336K2W1 A0A182QYX1 A0A084VBU8 A0A182JMA6 A0A182FNP8 A0A1B0D964 A0A182PBE8 A0A182N6V4 W5JJX4 A0A182MSA7 A0A2M4AJ10 A0A2M4AJ71 A0A182K0W3 A0A2M4AJE8 A0A182RBW8 A0A182I8H9 F5HIX4 F7IUV9 A0A182V9F1 U5ESJ0 A0A182WYH5 A0A1J1IGH9 A0A182LAD7 A0A182U109 A0A182VQD7 A0A1W4XEC0 A0A182XX98 A0A1W4XPF0 A0A1Y1NIS5 K7IP01 A0A1L8DJJ8 A0A1Q3F988 B0X5M3 A0A1S4FSQ1 A0A1S4FSV9 A0A310SK92 A0A232EVQ9 Q16QW1 J9HZJ7 E2A429 A0A026W7V4 A0A3L8D5I7 A0A2J7RH83 A0A023EWI7 A0A182G3Z5 A0A2J7RH63 U4UB39 A0A0L7RHE3 E2BWL5 A0A195ELC2 E9IKW0 K7ITA7 A0A0A9Y1F1 A0A146M613 A0A146LSJ3 A0A0A9WCY1 A0A146M5Z0 F4WL47 A0A0A9XGT7 D6WYS2 A0A232FDJ7 A0A1W4XPF5 A0A1W4XE01 A0A1W4XNL8 A0A3B0J912 B4NBZ8 A0A3B0JZF0 A0A3B0JDM7 B4L5W0 D0QWG5 A0A1A9WA16 A0A0R3P265 B4H345 A0A0R3NXK1 A0A0R3NXM4 A0A0R3NY26 B3MYY5 A0A195CIA3 A0A0Q9XF58 A0A0P8XH64 A0A0P9A6W1 A0A224XMI6 A0A1I8MEZ3 A0A151WM05 A0A0R3P2I7 Q29JK0 A0A1I8MEZ5 A0A1A9Z3Y6 O77266 A0A1I8PMJ8 A0A1B6CZA7
Pubmed
19121390
26227816
26354079
22118469
24438588
20920257
+ More
23761445 12364791 14747013 17210077 20966253 25244985 28004739 20075255 28648823 17510324 20798317 24508170 30249741 24945155 26483478 23537049 21282665 25401762 26823975 21719571 18362917 19820115 17994087 18057021 19859648 15632085 23185243 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867
23761445 12364791 14747013 17210077 20966253 25244985 28004739 20075255 28648823 17510324 20798317 24508170 30249741 24945155 26483478 23537049 21282665 25401762 26823975 21719571 18362917 19820115 17994087 18057021 19859648 15632085 23185243 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867
EMBL
BABH01038563
ODYU01000178
SOQ34528.1
JTDY01003600
KOB69285.1
NWSH01000035
+ More
PCG80421.1 KQ458756 KPJ05230.1 RSAL01000051 RVE50234.1 KQ460426 KPJ14799.1 AGBW02008320 OWR53754.1 UFQT01001992 SSX32339.1 UFQS01000056 UFQT01000056 SSW98741.1 SSX19127.1 AXCN02000309 ATLV01009527 KE524533 KFB35442.1 AJVK01004244 ADMH02001019 ETN64401.1 AXCM01006229 GGFK01007454 MBW40775.1 GGFK01007500 MBW40821.1 GGFK01007585 MBW40906.1 APCN01003033 AAAB01008805 EGK96235.1 EAA03876.4 GANO01003224 JAB56647.1 CVRI01000047 CRK98150.1 GEZM01005655 JAV96006.1 GFDF01007554 JAV06530.1 GFDL01010901 JAV24144.1 DS232386 EDS40960.1 KQ760478 OAD60226.1 NNAY01001962 OXU22439.1 CH477729 EAT36789.1 EJY57905.1 GL436526 EFN71795.1 KK107353 EZA52078.1 QOIP01000012 RLU15757.1 NEVH01003747 PNF40167.1 GAPW01000734 JAC12864.1 JXUM01041639 JXUM01041640 JXUM01041641 KQ561292 KXJ79092.1 PNF40168.1 KB632003 ERL87816.1 KQ414590 KOC70387.1 GL451131 EFN79971.1 KQ978747 KYN28679.1 GL764026 EFZ18689.1 AAZX01008304 AAZX01013357 GBHO01024438 GBHO01024437 GBHO01018138 GBHO01018137 GBHO01018131 GBRD01017410 JAG19166.1 JAG19167.1 JAG25466.1 JAG25467.1 JAG25473.1 JAG48417.1 GDHC01004177 JAQ14452.1 GDHC01008410 JAQ10219.1 GBHO01044355 GBHO01040884 GBHO01018136 GBHO01018134 GBHO01010453 GBRD01017411 JAF99248.1 JAG02720.1 JAG25468.1 JAG25470.1 JAG33151.1 JAG48416.1 GDHC01003465 JAQ15164.1 GL888207 EGI65010.1 GBHO01029525 GBHO01024435 GBHO01024432 GBHO01018135 GBHO01004291 JAG14079.1 JAG19169.1 JAG19172.1 JAG25469.1 JAG39313.1 KQ971357 EFA08401.2 NNAY01000418 OXU28548.1 OUUW01000003 SPP78744.1 CH964239 EDW82357.1 SPP78746.1 SPP78743.1 CH933811 EDW06569.1 KRG07020.1 KRG07022.1 FJ821027 ACN94653.1 CH379063 KRT05823.1 KRT05824.1 CH479205 EDW30738.1 KRT05822.1 KRT05825.1 KRT05820.1 CH902632 EDV32829.1 KQ977720 KYN00438.1 KRG07021.1 KPU74159.1 KPU74160.1 KPU74161.1 GFTR01007073 JAW09353.1 KQ982944 KYQ48863.1 KRT05821.1 EAL32300.3 AE014298 AAF45715.1 GEDC01018550 GEDC01016212 JAS18748.1 JAS21086.1
PCG80421.1 KQ458756 KPJ05230.1 RSAL01000051 RVE50234.1 KQ460426 KPJ14799.1 AGBW02008320 OWR53754.1 UFQT01001992 SSX32339.1 UFQS01000056 UFQT01000056 SSW98741.1 SSX19127.1 AXCN02000309 ATLV01009527 KE524533 KFB35442.1 AJVK01004244 ADMH02001019 ETN64401.1 AXCM01006229 GGFK01007454 MBW40775.1 GGFK01007500 MBW40821.1 GGFK01007585 MBW40906.1 APCN01003033 AAAB01008805 EGK96235.1 EAA03876.4 GANO01003224 JAB56647.1 CVRI01000047 CRK98150.1 GEZM01005655 JAV96006.1 GFDF01007554 JAV06530.1 GFDL01010901 JAV24144.1 DS232386 EDS40960.1 KQ760478 OAD60226.1 NNAY01001962 OXU22439.1 CH477729 EAT36789.1 EJY57905.1 GL436526 EFN71795.1 KK107353 EZA52078.1 QOIP01000012 RLU15757.1 NEVH01003747 PNF40167.1 GAPW01000734 JAC12864.1 JXUM01041639 JXUM01041640 JXUM01041641 KQ561292 KXJ79092.1 PNF40168.1 KB632003 ERL87816.1 KQ414590 KOC70387.1 GL451131 EFN79971.1 KQ978747 KYN28679.1 GL764026 EFZ18689.1 AAZX01008304 AAZX01013357 GBHO01024438 GBHO01024437 GBHO01018138 GBHO01018137 GBHO01018131 GBRD01017410 JAG19166.1 JAG19167.1 JAG25466.1 JAG25467.1 JAG25473.1 JAG48417.1 GDHC01004177 JAQ14452.1 GDHC01008410 JAQ10219.1 GBHO01044355 GBHO01040884 GBHO01018136 GBHO01018134 GBHO01010453 GBRD01017411 JAF99248.1 JAG02720.1 JAG25468.1 JAG25470.1 JAG33151.1 JAG48416.1 GDHC01003465 JAQ15164.1 GL888207 EGI65010.1 GBHO01029525 GBHO01024435 GBHO01024432 GBHO01018135 GBHO01004291 JAG14079.1 JAG19169.1 JAG19172.1 JAG25469.1 JAG39313.1 KQ971357 EFA08401.2 NNAY01000418 OXU28548.1 OUUW01000003 SPP78744.1 CH964239 EDW82357.1 SPP78746.1 SPP78743.1 CH933811 EDW06569.1 KRG07020.1 KRG07022.1 FJ821027 ACN94653.1 CH379063 KRT05823.1 KRT05824.1 CH479205 EDW30738.1 KRT05822.1 KRT05825.1 KRT05820.1 CH902632 EDV32829.1 KQ977720 KYN00438.1 KRG07021.1 KPU74159.1 KPU74160.1 KPU74161.1 GFTR01007073 JAW09353.1 KQ982944 KYQ48863.1 KRT05821.1 EAL32300.3 AE014298 AAF45715.1 GEDC01018550 GEDC01016212 JAS18748.1 JAS21086.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053268
UP000283053
UP000053240
+ More
UP000007151 UP000075886 UP000030765 UP000075880 UP000069272 UP000092462 UP000075885 UP000075884 UP000000673 UP000075883 UP000075881 UP000075900 UP000075840 UP000007062 UP000075903 UP000076407 UP000183832 UP000075882 UP000075902 UP000075920 UP000192223 UP000076408 UP000002358 UP000002320 UP000215335 UP000008820 UP000000311 UP000053097 UP000279307 UP000235965 UP000069940 UP000249989 UP000030742 UP000053825 UP000008237 UP000078492 UP000007755 UP000007266 UP000268350 UP000007798 UP000009192 UP000091820 UP000001819 UP000008744 UP000007801 UP000078542 UP000095301 UP000075809 UP000092445 UP000000803 UP000095300
UP000007151 UP000075886 UP000030765 UP000075880 UP000069272 UP000092462 UP000075885 UP000075884 UP000000673 UP000075883 UP000075881 UP000075900 UP000075840 UP000007062 UP000075903 UP000076407 UP000183832 UP000075882 UP000075902 UP000075920 UP000192223 UP000076408 UP000002358 UP000002320 UP000215335 UP000008820 UP000000311 UP000053097 UP000279307 UP000235965 UP000069940 UP000249989 UP000030742 UP000053825 UP000008237 UP000078492 UP000007755 UP000007266 UP000268350 UP000007798 UP000009192 UP000091820 UP000001819 UP000008744 UP000007801 UP000078542 UP000095301 UP000075809 UP000092445 UP000000803 UP000095300
Interpro
Gene 3D
ProteinModelPortal
H9JUA5
A0A2H1V112
A0A0L7L1H8
A0A2A4K867
A0A194QJZ1
A0A3S2NG59
+ More
A0A0N1IHV4 A0A212FJ44 A0A336MTC2 A0A336K2W1 A0A182QYX1 A0A084VBU8 A0A182JMA6 A0A182FNP8 A0A1B0D964 A0A182PBE8 A0A182N6V4 W5JJX4 A0A182MSA7 A0A2M4AJ10 A0A2M4AJ71 A0A182K0W3 A0A2M4AJE8 A0A182RBW8 A0A182I8H9 F5HIX4 F7IUV9 A0A182V9F1 U5ESJ0 A0A182WYH5 A0A1J1IGH9 A0A182LAD7 A0A182U109 A0A182VQD7 A0A1W4XEC0 A0A182XX98 A0A1W4XPF0 A0A1Y1NIS5 K7IP01 A0A1L8DJJ8 A0A1Q3F988 B0X5M3 A0A1S4FSQ1 A0A1S4FSV9 A0A310SK92 A0A232EVQ9 Q16QW1 J9HZJ7 E2A429 A0A026W7V4 A0A3L8D5I7 A0A2J7RH83 A0A023EWI7 A0A182G3Z5 A0A2J7RH63 U4UB39 A0A0L7RHE3 E2BWL5 A0A195ELC2 E9IKW0 K7ITA7 A0A0A9Y1F1 A0A146M613 A0A146LSJ3 A0A0A9WCY1 A0A146M5Z0 F4WL47 A0A0A9XGT7 D6WYS2 A0A232FDJ7 A0A1W4XPF5 A0A1W4XE01 A0A1W4XNL8 A0A3B0J912 B4NBZ8 A0A3B0JZF0 A0A3B0JDM7 B4L5W0 D0QWG5 A0A1A9WA16 A0A0R3P265 B4H345 A0A0R3NXK1 A0A0R3NXM4 A0A0R3NY26 B3MYY5 A0A195CIA3 A0A0Q9XF58 A0A0P8XH64 A0A0P9A6W1 A0A224XMI6 A0A1I8MEZ3 A0A151WM05 A0A0R3P2I7 Q29JK0 A0A1I8MEZ5 A0A1A9Z3Y6 O77266 A0A1I8PMJ8 A0A1B6CZA7
A0A0N1IHV4 A0A212FJ44 A0A336MTC2 A0A336K2W1 A0A182QYX1 A0A084VBU8 A0A182JMA6 A0A182FNP8 A0A1B0D964 A0A182PBE8 A0A182N6V4 W5JJX4 A0A182MSA7 A0A2M4AJ10 A0A2M4AJ71 A0A182K0W3 A0A2M4AJE8 A0A182RBW8 A0A182I8H9 F5HIX4 F7IUV9 A0A182V9F1 U5ESJ0 A0A182WYH5 A0A1J1IGH9 A0A182LAD7 A0A182U109 A0A182VQD7 A0A1W4XEC0 A0A182XX98 A0A1W4XPF0 A0A1Y1NIS5 K7IP01 A0A1L8DJJ8 A0A1Q3F988 B0X5M3 A0A1S4FSQ1 A0A1S4FSV9 A0A310SK92 A0A232EVQ9 Q16QW1 J9HZJ7 E2A429 A0A026W7V4 A0A3L8D5I7 A0A2J7RH83 A0A023EWI7 A0A182G3Z5 A0A2J7RH63 U4UB39 A0A0L7RHE3 E2BWL5 A0A195ELC2 E9IKW0 K7ITA7 A0A0A9Y1F1 A0A146M613 A0A146LSJ3 A0A0A9WCY1 A0A146M5Z0 F4WL47 A0A0A9XGT7 D6WYS2 A0A232FDJ7 A0A1W4XPF5 A0A1W4XE01 A0A1W4XNL8 A0A3B0J912 B4NBZ8 A0A3B0JZF0 A0A3B0JDM7 B4L5W0 D0QWG5 A0A1A9WA16 A0A0R3P265 B4H345 A0A0R3NXK1 A0A0R3NXM4 A0A0R3NY26 B3MYY5 A0A195CIA3 A0A0Q9XF58 A0A0P8XH64 A0A0P9A6W1 A0A224XMI6 A0A1I8MEZ3 A0A151WM05 A0A0R3P2I7 Q29JK0 A0A1I8MEZ5 A0A1A9Z3Y6 O77266 A0A1I8PMJ8 A0A1B6CZA7
PDB
4EMJ
E-value=3.14003e-30,
Score=330
Ontologies
GO
Topology
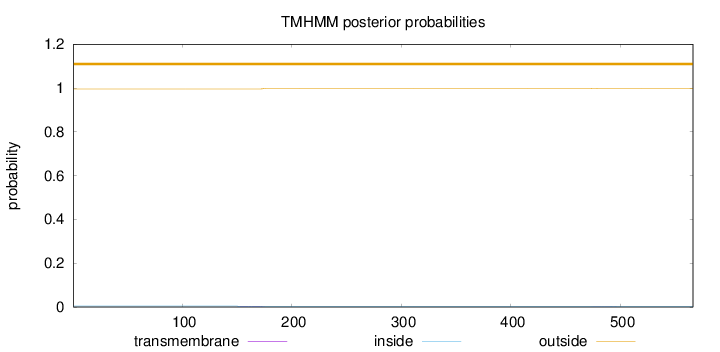
Length:
566
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.09048
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00470
outside
1 - 566
Population Genetic Test Statistics
Pi
18.583979
Theta
17.85539
Tajima's D
-0.643967
CLR
167.911683
CSRT
0.20313984300785
Interpretation
Uncertain
