Gene
KWMTBOMO09808 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013116
Annotation
chaperonin_subunit_6a_zeta_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.96
Sequence
CDS
ATGGCTGCCATTAGTTTATTAAATCCTAAAGCCGAGTTCGCTCGTGCTGCACAGGCACTTGCTGTAAATATATCCGCAGCTAAAGGAATTCAAGATGTAATGAAAACTAACCTTGGTCCGAAAGGCACGATGAAAATGTTGGTTTCTGGTGCTGGGGACATAAAGATCACCAAGGATGGCAATGTTTTGTTACATGAGATGCAAATCCAGCACCCTACAGCCTCACTGATTGCTCGAGCGTCAACCGCTCAAGATGATGCCACTGGAGATGGTACAACATCTACTGTCCTCCTCATTGGAGAGCTACTTAAACAGGCCGACATATTCATCAGTGAAGGCTTACATCCTAGGATCATTACTGAAGGCTTTGACATAGCTCGCAACAAGTCATTAGAAGTCTTGGAATCTATGAAGATTCCAATTGAGATTGCACGTGAGAATTTAGTTGATGTTGCCCGTACTTCACTCAAAACTAAGGTCCACCCAAGCTTGGCAGATGTGCTAACTGATGCTTGCGTTGATGCGGTACTCACCATTCGAACTCCAGGCAAACCTGTTGATCTGCACATGGTTGAAATTATGGAGATGAAACACAAGACGGCTACTGAAACTGTACTTGTAAAAGGTCTAGTAATGGATCATGGTGCACGTCACCCAGATATGCCAAAGCGTGTAGAAAATGCATACATTCTCACATGTAATGTGTCATTAGAGTATGAAAAGACAGAAGTAAATTCTGGCTTCTTCTACAAGTCTGCTGAGGATAGAGAAAAGCTTGTTGCTGCTGAAAGGGAATTTATTGATCAAAGAGTAAGAAAGATCGTTGCTCTCAAAAAGAAATTATGCGACGGTACTGACAAGACTTTCGTCGTTATCAACCAAAAAGGAATCGACCCCTTGTCACTGGATGCTTTTGCCAAAGAAGGTATTATTGGACTACGCAGAGCAAAACGGCGCAATATGGAACGTCTAGCCCTAGCGTGCGGGGGGTCCGCTGTTAACTCTGTCGATGATCTTACTGAAGACGCTTTAGGCTTTGCAGGCCTAGTCTACGAACACGTGCTCGGAGAAGACAAGTTTACCTTTGTTGAACAATGCAAGAATCCACAATCTGTCACCATCCTCATCAAGGGACCCAACAAGCATACGTTGACGCAAATCAAAGATGCAGTTCGTGACGGCCTCCGCGCTATTAATAACGCCATTGAAGATAAATGCGTCATACCCGGTGCTGCGGCCTTCGAAATCAAAGCTAACACGGAACTGCTGAAATATAAGGACACTGTTAAAGGCAAGGCCCGTCTTGGAATCCAAGCTTATGCCGAAGCTCTTTTGGTTATTCCTAAAACTCTGGCCGTTAACTCTGGTTACGATGCTCAAGATACTATAGTAAAGTTGCAGGAAGAGTCACGCCTTAATCCTGAGCCTATTGGCTTAGATTTGAGTACAGGTGAAGCATTCAAACCTACCGACTTAGGTATTTTGGACAACTATATCGTGAAGAAACAAATATTGAATTCCTGTTCAGTGATCGCAAGCAACCTCCTCCTTGTAGACGAGATTATGAGAGCAGGAATGAGCAGCCTCAAAGGATAA
Protein
MAAISLLNPKAEFARAAQALAVNISAAKGIQDVMKTNLGPKGTMKMLVSGAGDIKITKDGNVLLHEMQIQHPTASLIARASTAQDDATGDGTTSTVLLIGELLKQADIFISEGLHPRIITEGFDIARNKSLEVLESMKIPIEIARENLVDVARTSLKTKVHPSLADVLTDACVDAVLTIRTPGKPVDLHMVEIMEMKHKTATETVLVKGLVMDHGARHPDMPKRVENAYILTCNVSLEYEKTEVNSGFFYKSAEDREKLVAAEREFIDQRVRKIVALKKKLCDGTDKTFVVINQKGIDPLSLDAFAKEGIIGLRRAKRRNMERLALACGGSAVNSVDDLTEDALGFAGLVYEHVLGEDKFTFVEQCKNPQSVTILIKGPNKHTLTQIKDAVRDGLRAINNAIEDKCVIPGAAAFEIKANTELLKYKDTVKGKARLGIQAYAEALLVIPKTLAVNSGYDAQDTIVKLQEESRLNPEPIGLDLSTGEAFKPTDLGILDNYIVKKQILNSCSVIASNLLLVDEIMRAGMSSLKG
Summary
Similarity
Belongs to the TCP-1 chaperonin family.
Uniprot
Q2F6C3
A0A212FJ61
A0A2H1V2E8
A0A2A4K857
A0A3S2M367
S4PDW0
+ More
A0A0N1IEX5 A0A194QJZ8 A0A1L8EEA2 A0A1I8P4I6 A0A1L8EE95 W8C279 A0A0K8VQJ8 A0A034W9B6 T1PGT2 A0A0A1WXS6 A0A097P6I7 A0A0L0C137 A0A336LX66 A0A1W4XP63 A0A1Y1NDW3 D2A4I8 V5I9G4 A0A2J7PGY5 A0A067QHE4 A0A023ETN8 A0A023ETL9 Q174C6 N6TZB0 B3MXI0 A0A182P704 B4PX57 A0A0M4EJ31 W5J3C2 B3NTG6 A0A1W4V425 A0A2M3Z621 B5DLU0 A0A182VKE4 A0A182UJ87 A0A182KX61 A0A182XK15 Q7QDE6 A0A182HJX5 A0A0T6ASV4 A0A1B0C8A4 Q9VXQ5 U4USH2 V5RE81 A0A182QGR3 A0A182KDT6 A0A1A9XRB2 A0A182NKZ9 A0A1J1HTZ5 A0A1B0A037 A0A182VS76 U5EX39 A0A3B0KVS6 D3TMK9 A0A182YRK1 O96965 A0A182LWS2 B4NCT3 A0A084VTX8 A0A182R861 B4L2D4 B4JND6 A0A1Q3F2D5 A0A232EXZ9 B0W8W8 E2BN30 K7J5R2 A0A182F8I3 A0A2M4BJT7 A0A1J1HYD0 A0A2M4A6V7 A0A182IY84 A0A1B6I309 A0A1L8DUC7 B4GLB1 A0A1L8DUA0 B5DWX8 A0A1B6CFE0 E9J585 A0A3B0JU54 A0A0N0U3R7 A0A2A3EB60 V9IIN2 V4A0Q9 A0A310SE15 H9AWU2 A0A0J7L1D5 K1PXN5 A0A0L7R319 A0A151WMB8 E9FYC3 A0A151ICX0 A0A195E5K5 F4WPE3 A0A158NQZ3
A0A0N1IEX5 A0A194QJZ8 A0A1L8EEA2 A0A1I8P4I6 A0A1L8EE95 W8C279 A0A0K8VQJ8 A0A034W9B6 T1PGT2 A0A0A1WXS6 A0A097P6I7 A0A0L0C137 A0A336LX66 A0A1W4XP63 A0A1Y1NDW3 D2A4I8 V5I9G4 A0A2J7PGY5 A0A067QHE4 A0A023ETN8 A0A023ETL9 Q174C6 N6TZB0 B3MXI0 A0A182P704 B4PX57 A0A0M4EJ31 W5J3C2 B3NTG6 A0A1W4V425 A0A2M3Z621 B5DLU0 A0A182VKE4 A0A182UJ87 A0A182KX61 A0A182XK15 Q7QDE6 A0A182HJX5 A0A0T6ASV4 A0A1B0C8A4 Q9VXQ5 U4USH2 V5RE81 A0A182QGR3 A0A182KDT6 A0A1A9XRB2 A0A182NKZ9 A0A1J1HTZ5 A0A1B0A037 A0A182VS76 U5EX39 A0A3B0KVS6 D3TMK9 A0A182YRK1 O96965 A0A182LWS2 B4NCT3 A0A084VTX8 A0A182R861 B4L2D4 B4JND6 A0A1Q3F2D5 A0A232EXZ9 B0W8W8 E2BN30 K7J5R2 A0A182F8I3 A0A2M4BJT7 A0A1J1HYD0 A0A2M4A6V7 A0A182IY84 A0A1B6I309 A0A1L8DUC7 B4GLB1 A0A1L8DUA0 B5DWX8 A0A1B6CFE0 E9J585 A0A3B0JU54 A0A0N0U3R7 A0A2A3EB60 V9IIN2 V4A0Q9 A0A310SE15 H9AWU2 A0A0J7L1D5 K1PXN5 A0A0L7R319 A0A151WMB8 E9FYC3 A0A151ICX0 A0A195E5K5 F4WPE3 A0A158NQZ3
Pubmed
19121390
22118469
23622113
26354079
24495485
25348373
+ More
25830018 25315136 26108605 28004739 18362917 19820115 24845553 24945155 26483478 17510324 23537049 17994087 17550304 20920257 23761445 15632085 20966253 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20353571 25244985 18057021 24438588 28648823 20798317 20075255 21282665 23254933 22992520 21292972 21719571 21347285
25830018 25315136 26108605 28004739 18362917 19820115 24845553 24945155 26483478 17510324 23537049 17994087 17550304 20920257 23761445 15632085 20966253 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20353571 25244985 18057021 24438588 28648823 20798317 20075255 21282665 23254933 22992520 21292972 21719571 21347285
EMBL
BABH01038575
DQ311149
ABD36094.1
AGBW02008320
OWR53749.1
ODYU01000178
+ More
SOQ34532.1 NWSH01000035 PCG80417.1 RSAL01000051 RVE50238.1 GAIX01003476 JAA89084.1 KQ460426 KPJ14804.1 KQ458756 KPJ05235.1 GFDG01001762 JAV17037.1 GFDG01001779 JAV17020.1 GAMC01003247 JAC03309.1 GDHF01011155 JAI41159.1 GAKP01007658 JAC51294.1 KA647093 AFP61722.1 GBXI01011074 JAD03218.1 KM215143 AIU39616.1 JRES01001143 KNC25174.1 UFQT01000271 SSX22642.1 GEZM01008450 JAV94775.1 KQ971344 EFA05810.1 GALX01002980 JAB65486.1 NEVH01025161 PNF15588.1 KK853387 KDR07944.1 GAPW01000900 JAC12698.1 JXUM01245109 GAPW01000908 KQ636705 JAC12690.1 KXJ62410.1 CH477414 EAT41398.1 APGK01055164 KB741261 ENN71572.1 CH902630 EDV38445.1 CM000162 EDX01820.1 CP012528 ALC49054.1 ADMH02002206 ETN57823.1 CH954180 EDV46970.1 GGFM01003194 MBW23945.1 CH379064 EDY72486.1 AAAB01008851 EAA07393.4 APCN01002215 LJIG01022882 KRT78240.1 AJWK01000296 AE014298 AY051470 AAF48503.1 AAK92894.1 KB632344 ERL93050.1 KF621135 AHB33471.1 AXCN02000100 CVRI01000021 CRK91543.1 GANO01001137 JAB58734.1 OUUW01000042 SPP89976.1 CCAG010015881 EZ422661 ADD18937.1 Y18419 CH940664 CAA77160.1 EDW63022.1 AXCM01003454 CH964239 EDW82642.1 ATLV01016551 KE525093 KFB41422.1 CH933810 EDW07795.1 CH916371 EDV92229.1 GFDL01013320 JAV21725.1 NNAY01001696 OXU23195.1 DS231860 EDS39347.1 GL449382 EFN82934.1 GGFJ01004198 MBW53339.1 CRK91542.1 GGFK01003037 MBW36358.1 GECU01026402 JAS81304.1 GFDF01004064 JAV10020.1 CH479185 EDW38335.1 GFDF01004065 JAV10019.1 CM000070 EDY68580.1 GEDC01025111 JAS12187.1 GL768124 EFZ12013.1 OUUW01000008 SPP83932.1 KQ435885 KOX69602.1 KZ288295 PBC28967.1 JR047758 AEY60497.1 KB201305 ESO97378.1 KQ760801 OAD58980.1 JN899240 AFC98245.1 LBMM01001295 KMQ96562.1 JH815665 EKC21190.1 KQ414663 KOC65272.1 KQ982944 KYQ48983.1 GL732527 EFX87506.1 KQ978052 KYM97788.1 KQ979608 KYN20443.1 GL888243 EGI64038.1 ADTU01023710
SOQ34532.1 NWSH01000035 PCG80417.1 RSAL01000051 RVE50238.1 GAIX01003476 JAA89084.1 KQ460426 KPJ14804.1 KQ458756 KPJ05235.1 GFDG01001762 JAV17037.1 GFDG01001779 JAV17020.1 GAMC01003247 JAC03309.1 GDHF01011155 JAI41159.1 GAKP01007658 JAC51294.1 KA647093 AFP61722.1 GBXI01011074 JAD03218.1 KM215143 AIU39616.1 JRES01001143 KNC25174.1 UFQT01000271 SSX22642.1 GEZM01008450 JAV94775.1 KQ971344 EFA05810.1 GALX01002980 JAB65486.1 NEVH01025161 PNF15588.1 KK853387 KDR07944.1 GAPW01000900 JAC12698.1 JXUM01245109 GAPW01000908 KQ636705 JAC12690.1 KXJ62410.1 CH477414 EAT41398.1 APGK01055164 KB741261 ENN71572.1 CH902630 EDV38445.1 CM000162 EDX01820.1 CP012528 ALC49054.1 ADMH02002206 ETN57823.1 CH954180 EDV46970.1 GGFM01003194 MBW23945.1 CH379064 EDY72486.1 AAAB01008851 EAA07393.4 APCN01002215 LJIG01022882 KRT78240.1 AJWK01000296 AE014298 AY051470 AAF48503.1 AAK92894.1 KB632344 ERL93050.1 KF621135 AHB33471.1 AXCN02000100 CVRI01000021 CRK91543.1 GANO01001137 JAB58734.1 OUUW01000042 SPP89976.1 CCAG010015881 EZ422661 ADD18937.1 Y18419 CH940664 CAA77160.1 EDW63022.1 AXCM01003454 CH964239 EDW82642.1 ATLV01016551 KE525093 KFB41422.1 CH933810 EDW07795.1 CH916371 EDV92229.1 GFDL01013320 JAV21725.1 NNAY01001696 OXU23195.1 DS231860 EDS39347.1 GL449382 EFN82934.1 GGFJ01004198 MBW53339.1 CRK91542.1 GGFK01003037 MBW36358.1 GECU01026402 JAS81304.1 GFDF01004064 JAV10020.1 CH479185 EDW38335.1 GFDF01004065 JAV10019.1 CM000070 EDY68580.1 GEDC01025111 JAS12187.1 GL768124 EFZ12013.1 OUUW01000008 SPP83932.1 KQ435885 KOX69602.1 KZ288295 PBC28967.1 JR047758 AEY60497.1 KB201305 ESO97378.1 KQ760801 OAD58980.1 JN899240 AFC98245.1 LBMM01001295 KMQ96562.1 JH815665 EKC21190.1 KQ414663 KOC65272.1 KQ982944 KYQ48983.1 GL732527 EFX87506.1 KQ978052 KYM97788.1 KQ979608 KYN20443.1 GL888243 EGI64038.1 ADTU01023710
Proteomes
UP000005204
UP000007151
UP000218220
UP000283053
UP000053240
UP000053268
+ More
UP000095300 UP000095301 UP000037069 UP000192223 UP000007266 UP000235965 UP000027135 UP000069940 UP000249989 UP000008820 UP000019118 UP000007801 UP000075885 UP000002282 UP000092553 UP000000673 UP000008711 UP000192221 UP000001819 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000092461 UP000000803 UP000030742 UP000075886 UP000075881 UP000092443 UP000075884 UP000183832 UP000092445 UP000075920 UP000268350 UP000092444 UP000076408 UP000008792 UP000075883 UP000007798 UP000030765 UP000075900 UP000009192 UP000001070 UP000215335 UP000002320 UP000008237 UP000002358 UP000069272 UP000075880 UP000008744 UP000053105 UP000242457 UP000030746 UP000036403 UP000005408 UP000053825 UP000075809 UP000000305 UP000078542 UP000078492 UP000007755 UP000005205
UP000095300 UP000095301 UP000037069 UP000192223 UP000007266 UP000235965 UP000027135 UP000069940 UP000249989 UP000008820 UP000019118 UP000007801 UP000075885 UP000002282 UP000092553 UP000000673 UP000008711 UP000192221 UP000001819 UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000092461 UP000000803 UP000030742 UP000075886 UP000075881 UP000092443 UP000075884 UP000183832 UP000092445 UP000075920 UP000268350 UP000092444 UP000076408 UP000008792 UP000075883 UP000007798 UP000030765 UP000075900 UP000009192 UP000001070 UP000215335 UP000002320 UP000008237 UP000002358 UP000069272 UP000075880 UP000008744 UP000053105 UP000242457 UP000030746 UP000036403 UP000005408 UP000053825 UP000075809 UP000000305 UP000078542 UP000078492 UP000007755 UP000005205
PRIDE
Pfam
PF00118 Cpn60_TCP1
Interpro
Gene 3D
ProteinModelPortal
Q2F6C3
A0A212FJ61
A0A2H1V2E8
A0A2A4K857
A0A3S2M367
S4PDW0
+ More
A0A0N1IEX5 A0A194QJZ8 A0A1L8EEA2 A0A1I8P4I6 A0A1L8EE95 W8C279 A0A0K8VQJ8 A0A034W9B6 T1PGT2 A0A0A1WXS6 A0A097P6I7 A0A0L0C137 A0A336LX66 A0A1W4XP63 A0A1Y1NDW3 D2A4I8 V5I9G4 A0A2J7PGY5 A0A067QHE4 A0A023ETN8 A0A023ETL9 Q174C6 N6TZB0 B3MXI0 A0A182P704 B4PX57 A0A0M4EJ31 W5J3C2 B3NTG6 A0A1W4V425 A0A2M3Z621 B5DLU0 A0A182VKE4 A0A182UJ87 A0A182KX61 A0A182XK15 Q7QDE6 A0A182HJX5 A0A0T6ASV4 A0A1B0C8A4 Q9VXQ5 U4USH2 V5RE81 A0A182QGR3 A0A182KDT6 A0A1A9XRB2 A0A182NKZ9 A0A1J1HTZ5 A0A1B0A037 A0A182VS76 U5EX39 A0A3B0KVS6 D3TMK9 A0A182YRK1 O96965 A0A182LWS2 B4NCT3 A0A084VTX8 A0A182R861 B4L2D4 B4JND6 A0A1Q3F2D5 A0A232EXZ9 B0W8W8 E2BN30 K7J5R2 A0A182F8I3 A0A2M4BJT7 A0A1J1HYD0 A0A2M4A6V7 A0A182IY84 A0A1B6I309 A0A1L8DUC7 B4GLB1 A0A1L8DUA0 B5DWX8 A0A1B6CFE0 E9J585 A0A3B0JU54 A0A0N0U3R7 A0A2A3EB60 V9IIN2 V4A0Q9 A0A310SE15 H9AWU2 A0A0J7L1D5 K1PXN5 A0A0L7R319 A0A151WMB8 E9FYC3 A0A151ICX0 A0A195E5K5 F4WPE3 A0A158NQZ3
A0A0N1IEX5 A0A194QJZ8 A0A1L8EEA2 A0A1I8P4I6 A0A1L8EE95 W8C279 A0A0K8VQJ8 A0A034W9B6 T1PGT2 A0A0A1WXS6 A0A097P6I7 A0A0L0C137 A0A336LX66 A0A1W4XP63 A0A1Y1NDW3 D2A4I8 V5I9G4 A0A2J7PGY5 A0A067QHE4 A0A023ETN8 A0A023ETL9 Q174C6 N6TZB0 B3MXI0 A0A182P704 B4PX57 A0A0M4EJ31 W5J3C2 B3NTG6 A0A1W4V425 A0A2M3Z621 B5DLU0 A0A182VKE4 A0A182UJ87 A0A182KX61 A0A182XK15 Q7QDE6 A0A182HJX5 A0A0T6ASV4 A0A1B0C8A4 Q9VXQ5 U4USH2 V5RE81 A0A182QGR3 A0A182KDT6 A0A1A9XRB2 A0A182NKZ9 A0A1J1HTZ5 A0A1B0A037 A0A182VS76 U5EX39 A0A3B0KVS6 D3TMK9 A0A182YRK1 O96965 A0A182LWS2 B4NCT3 A0A084VTX8 A0A182R861 B4L2D4 B4JND6 A0A1Q3F2D5 A0A232EXZ9 B0W8W8 E2BN30 K7J5R2 A0A182F8I3 A0A2M4BJT7 A0A1J1HYD0 A0A2M4A6V7 A0A182IY84 A0A1B6I309 A0A1L8DUC7 B4GLB1 A0A1L8DUA0 B5DWX8 A0A1B6CFE0 E9J585 A0A3B0JU54 A0A0N0U3R7 A0A2A3EB60 V9IIN2 V4A0Q9 A0A310SE15 H9AWU2 A0A0J7L1D5 K1PXN5 A0A0L7R319 A0A151WMB8 E9FYC3 A0A151ICX0 A0A195E5K5 F4WPE3 A0A158NQZ3
PDB
4B2T
E-value=0,
Score=1899
Ontologies
GO
Topology
Subcellular location
Cytoplasm
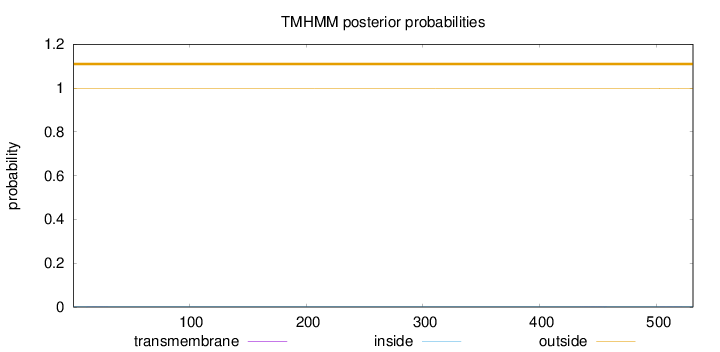
Length:
531
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01865
Exp number, first 60 AAs:
0.0027
Total prob of N-in:
0.00085
outside
1 - 531
Population Genetic Test Statistics
Pi
25.736773
Theta
204.082191
Tajima's D
0.502821
CLR
0.007264
CSRT
0.521173941302935
Interpretation
Uncertain
