Pre Gene Modal
BGIBMGA013148
Annotation
diphthamide_synthesis_protein_[Bombyx_mori]
Full name
2-(3-amino-3-carboxypropyl)histidine synthase subunit 1
Location in the cell
Cytoplasmic Reliability : 1.228
Sequence
CDS
ATGGAAGATTTTACCAATAATCCTGGTATAGTTGTTATACGTGCGAAACCGGAAGGACAAAGAAAAACTTTTAAACCCAAAGTTAGATCTTTAAATAAAATACCTGATGATTTGTTGAATGATCCCCTGCTAAACAGGGCTTGCGCGGCTTTACCAGAAAATTATAATTTCGAAATACACAAGACGATATGGAGAATACGGTCTACAGCGGCAAAACGCGTGGCCCTACAGATGCCTGAAGGTCTAACTATGTTTGCAACGACCCTCTGTGACATAATAGAAACATTCACCGAAGCCGATACTGTGATAATGGGAGATGTGACATACGGAGCCTGCTGTATAGATGACTTCACTGCTGTGGCACTCGGTGTAGACCTCCTTGTGCACTATGGACACTCTTGCCTAATTCCCATAGATCAAACTAGTAGTATTAAGGTTCTGTACATATTTGTGGATATAAAAATTGATCCGTCACATTTCATAGAAACAATAAAAGTAAACTTTCCATCTAAAACTCATTTAGCATTAGTTAGCACAATTCAATTCGTAACCACTCTACATTCTGTAGCTAAGAATCTTCGTATTGAAGAATACATGGTTACAGTGCCACAAACTAAACCCCTTTCACCAGGAGAGATACTAGGATGCACTGCTCCTAAATTACAAGCAGATGCAATTGTTTATTTAGGAGATGGAAGATTTCATTTAGAATCTATAATGATAGCGAATCCTACAATAAGTGCCTACAAATATGATCCATATGATAAAAAATTTACAAAAGAAGAATATGATCATGTTCTCATGAAGGCCAACAGACAGAAGCAAGTGAAGACTGCGGAGTCTGCAAGTAATTTTGGCCTTATTCTTGGCACTCTTGGAAGACAGGGCAGCACAAAAGTGTTAGCAAATTTAGAAAAGCAATTATCAAACTCTGATAAACATTATGTAAAGATATTGCTATCAGAGATTTTTCCTAGCAAACTTGCCTTATTTGGACTGGATGCATTTGTTCAGTGA
Protein
MEDFTNNPGIVVIRAKPEGQRKTFKPKVRSLNKIPDDLLNDPLLNRACAALPENYNFEIHKTIWRIRSTAAKRVALQMPEGLTMFATTLCDIIETFTEADTVIMGDVTYGACCIDDFTAVALGVDLLVHYGHSCLIPIDQTSSIKVLYIFVDIKIDPSHFIETIKVNFPSKTHLALVSTIQFVTTLHSVAKNLRIEEYMVTVPQTKPLSPGEILGCTAPKLQADAIVYLGDGRFHLESIMIANPTISAYKYDPYDKKFTKEEYDHVLMKANRQKQVKTAESASNFGLILGTLGRQGSTKVLANLEKQLSNSDKHYVKILLSEIFPSKLALFGLDAFVQ
Summary
Description
Required for the first step of diphthamide biosynthesis, the transfer of 3-amino-3-carboxypropyl from S-adenosyl-L-methionine to a histidine residue. Diphthamide is a post-translational modification of histidine which occurs in elongation factor 2.
Catalytic Activity
L-histidyl-[translation elongation factor 2] + S-adenosyl-L-methionine = 2-[(3S)-amino-3-carboxypropyl]-L-histidyl-[translation elongation factor 2] + H(+) + S-methyl-5'-thioadenosine
Similarity
Belongs to the DPH1/DPH2 family. DPH1 subfamily.
Belongs to the DPH1/DPH2 family.
Belongs to the DPH1/DPH2 family.
Feature
chain 2-(3-amino-3-carboxypropyl)histidine synthase subunit 1
Uniprot
H9JUD5
Q2F5S3
A0A0L7LAW6
S4PV65
A0A2H1VL98
A0A212FJ23
+ More
A0A1B0D580 A0A1L8DRK0 A0A1L8DRV4 A0A1J1J1X5 A0A182JIQ6 A0A1B0C9D6 A0A023ESE3 A0A182QRW7 A0A067QWF0 A0A182Y5X8 A0A182U1I3 A0A182SX18 A0A182N9L6 A0A182LUX9 A0A2J7QGS8 A0A182KDX2 A0A2M4ANX9 A0A182LP79 A0A2M4AP09 A0A182WEV6 A0A182RD29 A0A1B6I3C7 A0A2S2R5G8 A0A182FC28 A0A2M3Z7Q1 A0A182VG11 A0A2M3ZI03 A0A2M4BM58 A0A2M4CSS7 A0A2M4CSJ5 A0A182HNC2 A0A1B6FQL0 A0A2M4BM35 A0A182XC23 A0A336K702 A0A2R7VZ90 A0A034VQ42 A0A0K8UBF9 A0A182PL67 A0A0A1WEP6 A0A1Q3F6G1 D6WDC5 J9JPJ3 W8C3A4 A0A069DSX3 A0A0V0GCI1 A0A023FAB1 B0WLD0 B4IZC0 A0A0P4VV67 R4FKY8 A0A1B6D814 E9H287 K7IMS9 A0A0P5ZG97 A0A1A9WH61 B4N4M0 A0A162CC92 A0A0P6A778 A0A0P4YDQ0 A0A1I8P1D5 A0A0P4XZ24 A0A0P5U319 A0A0M3QWZ7 A0A0C9R5G0 B4LCR0 B4HAH3 A0A0P6J0J1 A0A0Q9WKC7 Q2LZ76 A0A0Q9WSH7 A0A0L0CEC1 A0A146LEQ0 B4QQ98 T1PK69 A0A0A9Y073 A0A1I8MLV5 A0A0A9XYR1 B4HEX3 Q9VTM2 B3NGZ2 A0A1W4W6K0 B4PFM8 A0A0P5DLK9 B4KUS5 E0VAN8 A0A3B0J8Y3 A0A3Q3G744 A0A0P5DFU1 A0A0P5D7I4 A0A2D0SW18 A0A0P5ZRK5 A0A1A8DR56 A0A3B0J9X1
A0A1B0D580 A0A1L8DRK0 A0A1L8DRV4 A0A1J1J1X5 A0A182JIQ6 A0A1B0C9D6 A0A023ESE3 A0A182QRW7 A0A067QWF0 A0A182Y5X8 A0A182U1I3 A0A182SX18 A0A182N9L6 A0A182LUX9 A0A2J7QGS8 A0A182KDX2 A0A2M4ANX9 A0A182LP79 A0A2M4AP09 A0A182WEV6 A0A182RD29 A0A1B6I3C7 A0A2S2R5G8 A0A182FC28 A0A2M3Z7Q1 A0A182VG11 A0A2M3ZI03 A0A2M4BM58 A0A2M4CSS7 A0A2M4CSJ5 A0A182HNC2 A0A1B6FQL0 A0A2M4BM35 A0A182XC23 A0A336K702 A0A2R7VZ90 A0A034VQ42 A0A0K8UBF9 A0A182PL67 A0A0A1WEP6 A0A1Q3F6G1 D6WDC5 J9JPJ3 W8C3A4 A0A069DSX3 A0A0V0GCI1 A0A023FAB1 B0WLD0 B4IZC0 A0A0P4VV67 R4FKY8 A0A1B6D814 E9H287 K7IMS9 A0A0P5ZG97 A0A1A9WH61 B4N4M0 A0A162CC92 A0A0P6A778 A0A0P4YDQ0 A0A1I8P1D5 A0A0P4XZ24 A0A0P5U319 A0A0M3QWZ7 A0A0C9R5G0 B4LCR0 B4HAH3 A0A0P6J0J1 A0A0Q9WKC7 Q2LZ76 A0A0Q9WSH7 A0A0L0CEC1 A0A146LEQ0 B4QQ98 T1PK69 A0A0A9Y073 A0A1I8MLV5 A0A0A9XYR1 B4HEX3 Q9VTM2 B3NGZ2 A0A1W4W6K0 B4PFM8 A0A0P5DLK9 B4KUS5 E0VAN8 A0A3B0J8Y3 A0A3Q3G744 A0A0P5DFU1 A0A0P5D7I4 A0A2D0SW18 A0A0P5ZRK5 A0A1A8DR56 A0A3B0J9X1
EC Number
2.5.1.108
Pubmed
19121390
26227816
23622113
22118469
24945155
24845553
+ More
25244985 20966253 25348373 25830018 18362917 19820115 24495485 26334808 25474469 17994087 27129103 21292972 20075255 18057021 15632085 23185243 26108605 26823975 22936249 25401762 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20566863
25244985 20966253 25348373 25830018 18362917 19820115 24495485 26334808 25474469 17994087 27129103 21292972 20075255 18057021 15632085 23185243 26108605 26823975 22936249 25401762 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20566863
EMBL
BABH01038580
DQ311350
ABD36294.1
JTDY01001999
KOB72361.1
GAIX01009078
+ More
JAA83482.1 ODYU01002923 SOQ41024.1 AGBW02008320 OWR53743.1 AJVK01003397 AJVK01003398 AJVK01003399 AJVK01003400 GFDF01004976 JAV09108.1 GFDF01004977 JAV09107.1 CVRI01000063 CRL04857.1 AJWK01002233 GAPW01001431 JAC12167.1 AXCN02002036 KK852879 KDR14464.1 AXCM01003863 NEVH01014359 PNF27788.1 GGFK01009169 MBW42490.1 GGFK01009203 MBW42524.1 GECU01026258 JAS81448.1 GGMS01016032 MBY85235.1 GGFM01003805 MBW24556.1 GGFM01007344 MBW28095.1 GGFJ01004994 MBW54135.1 GGFL01004137 MBW68315.1 GGFL01004136 MBW68314.1 APCN01001904 GECZ01017340 JAS52429.1 GGFJ01004996 MBW54137.1 UFQS01000131 UFQT01000131 SSX00237.1 SSX20617.1 KK854186 PTY12782.1 GAKP01015284 JAC43668.1 GDHF01028335 JAI23979.1 GBXI01017126 JAC97165.1 GFDL01011890 JAV23155.1 KQ971321 EEZ99477.1 ABLF02027856 GAMC01010247 GAMC01010246 JAB96309.1 GBGD01001696 JAC87193.1 GECL01000515 JAP05609.1 GBBI01000849 JAC17863.1 DS231984 EDS30423.1 CH916366 EDV96675.1 GDKW01001897 JAI54698.1 ACPB03004220 GAHY01001643 JAA75867.1 GEDC01015470 JAS21828.1 GL732585 EFX74125.1 GDIP01045770 JAM57945.1 CH964095 EDW79094.1 LRGB01001005 KZS14052.1 GDIP01034195 JAM69520.1 GDIP01238643 JAI84758.1 GDIP01238642 JAI84759.1 GDIP01120701 JAL83013.1 CP012525 ALC45053.1 GBYB01002066 JAG71833.1 CH940647 EDW70952.1 KRF85206.1 CH479240 EDW37571.1 GDIQ01024761 JAN69976.1 KRF85209.1 CH379069 EAL29632.1 KRF85207.1 JRES01000502 KNC30600.1 GDHC01013629 JAQ05000.1 CM000363 CM002912 EDX10116.1 KMY99046.1 KA648358 AFP62987.1 GBHO01019096 GBHO01019095 GBHO01019094 GBHO01004433 GBRD01002551 GDHC01019971 GDHC01018994 GDHC01014745 JAG24508.1 JAG24509.1 JAG24510.1 JAG39171.1 JAG63270.1 JAP98657.1 JAP99634.1 JAQ03884.1 GBHO01019097 GBHO01019093 JAG24507.1 JAG24511.1 CH480815 EDW41142.1 AE014296 AY069411 AAF50025.1 AAL39556.1 CH954178 EDV51449.1 CM000159 EDW94177.1 GDIP01154550 JAJ68852.1 CH933809 EDW19331.1 DS235012 EEB10444.1 OUUW01000002 SPP76773.1 GDIP01156899 JAJ66503.1 GDIP01161950 JAJ61452.1 GDIP01041061 JAM62654.1 HAEA01007573 SBQ36053.1 SPP76772.1
JAA83482.1 ODYU01002923 SOQ41024.1 AGBW02008320 OWR53743.1 AJVK01003397 AJVK01003398 AJVK01003399 AJVK01003400 GFDF01004976 JAV09108.1 GFDF01004977 JAV09107.1 CVRI01000063 CRL04857.1 AJWK01002233 GAPW01001431 JAC12167.1 AXCN02002036 KK852879 KDR14464.1 AXCM01003863 NEVH01014359 PNF27788.1 GGFK01009169 MBW42490.1 GGFK01009203 MBW42524.1 GECU01026258 JAS81448.1 GGMS01016032 MBY85235.1 GGFM01003805 MBW24556.1 GGFM01007344 MBW28095.1 GGFJ01004994 MBW54135.1 GGFL01004137 MBW68315.1 GGFL01004136 MBW68314.1 APCN01001904 GECZ01017340 JAS52429.1 GGFJ01004996 MBW54137.1 UFQS01000131 UFQT01000131 SSX00237.1 SSX20617.1 KK854186 PTY12782.1 GAKP01015284 JAC43668.1 GDHF01028335 JAI23979.1 GBXI01017126 JAC97165.1 GFDL01011890 JAV23155.1 KQ971321 EEZ99477.1 ABLF02027856 GAMC01010247 GAMC01010246 JAB96309.1 GBGD01001696 JAC87193.1 GECL01000515 JAP05609.1 GBBI01000849 JAC17863.1 DS231984 EDS30423.1 CH916366 EDV96675.1 GDKW01001897 JAI54698.1 ACPB03004220 GAHY01001643 JAA75867.1 GEDC01015470 JAS21828.1 GL732585 EFX74125.1 GDIP01045770 JAM57945.1 CH964095 EDW79094.1 LRGB01001005 KZS14052.1 GDIP01034195 JAM69520.1 GDIP01238643 JAI84758.1 GDIP01238642 JAI84759.1 GDIP01120701 JAL83013.1 CP012525 ALC45053.1 GBYB01002066 JAG71833.1 CH940647 EDW70952.1 KRF85206.1 CH479240 EDW37571.1 GDIQ01024761 JAN69976.1 KRF85209.1 CH379069 EAL29632.1 KRF85207.1 JRES01000502 KNC30600.1 GDHC01013629 JAQ05000.1 CM000363 CM002912 EDX10116.1 KMY99046.1 KA648358 AFP62987.1 GBHO01019096 GBHO01019095 GBHO01019094 GBHO01004433 GBRD01002551 GDHC01019971 GDHC01018994 GDHC01014745 JAG24508.1 JAG24509.1 JAG24510.1 JAG39171.1 JAG63270.1 JAP98657.1 JAP99634.1 JAQ03884.1 GBHO01019097 GBHO01019093 JAG24507.1 JAG24511.1 CH480815 EDW41142.1 AE014296 AY069411 AAF50025.1 AAL39556.1 CH954178 EDV51449.1 CM000159 EDW94177.1 GDIP01154550 JAJ68852.1 CH933809 EDW19331.1 DS235012 EEB10444.1 OUUW01000002 SPP76773.1 GDIP01156899 JAJ66503.1 GDIP01161950 JAJ61452.1 GDIP01041061 JAM62654.1 HAEA01007573 SBQ36053.1 SPP76772.1
Proteomes
UP000005204
UP000037510
UP000007151
UP000092462
UP000183832
UP000075880
+ More
UP000092461 UP000075886 UP000027135 UP000076408 UP000075902 UP000075901 UP000075884 UP000075883 UP000235965 UP000075881 UP000075882 UP000075920 UP000075900 UP000069272 UP000075903 UP000075840 UP000076407 UP000075885 UP000007266 UP000007819 UP000002320 UP000001070 UP000015103 UP000000305 UP000002358 UP000091820 UP000007798 UP000076858 UP000095300 UP000092553 UP000008792 UP000008744 UP000001819 UP000037069 UP000000304 UP000095301 UP000001292 UP000000803 UP000008711 UP000192221 UP000002282 UP000009192 UP000009046 UP000268350 UP000261660 UP000221080
UP000092461 UP000075886 UP000027135 UP000076408 UP000075902 UP000075901 UP000075884 UP000075883 UP000235965 UP000075881 UP000075882 UP000075920 UP000075900 UP000069272 UP000075903 UP000075840 UP000076407 UP000075885 UP000007266 UP000007819 UP000002320 UP000001070 UP000015103 UP000000305 UP000002358 UP000091820 UP000007798 UP000076858 UP000095300 UP000092553 UP000008792 UP000008744 UP000001819 UP000037069 UP000000304 UP000095301 UP000001292 UP000000803 UP000008711 UP000192221 UP000002282 UP000009192 UP000009046 UP000268350 UP000261660 UP000221080
Pfam
PF01866 Diphthamide_syn
Interpro
Gene 3D
ProteinModelPortal
H9JUD5
Q2F5S3
A0A0L7LAW6
S4PV65
A0A2H1VL98
A0A212FJ23
+ More
A0A1B0D580 A0A1L8DRK0 A0A1L8DRV4 A0A1J1J1X5 A0A182JIQ6 A0A1B0C9D6 A0A023ESE3 A0A182QRW7 A0A067QWF0 A0A182Y5X8 A0A182U1I3 A0A182SX18 A0A182N9L6 A0A182LUX9 A0A2J7QGS8 A0A182KDX2 A0A2M4ANX9 A0A182LP79 A0A2M4AP09 A0A182WEV6 A0A182RD29 A0A1B6I3C7 A0A2S2R5G8 A0A182FC28 A0A2M3Z7Q1 A0A182VG11 A0A2M3ZI03 A0A2M4BM58 A0A2M4CSS7 A0A2M4CSJ5 A0A182HNC2 A0A1B6FQL0 A0A2M4BM35 A0A182XC23 A0A336K702 A0A2R7VZ90 A0A034VQ42 A0A0K8UBF9 A0A182PL67 A0A0A1WEP6 A0A1Q3F6G1 D6WDC5 J9JPJ3 W8C3A4 A0A069DSX3 A0A0V0GCI1 A0A023FAB1 B0WLD0 B4IZC0 A0A0P4VV67 R4FKY8 A0A1B6D814 E9H287 K7IMS9 A0A0P5ZG97 A0A1A9WH61 B4N4M0 A0A162CC92 A0A0P6A778 A0A0P4YDQ0 A0A1I8P1D5 A0A0P4XZ24 A0A0P5U319 A0A0M3QWZ7 A0A0C9R5G0 B4LCR0 B4HAH3 A0A0P6J0J1 A0A0Q9WKC7 Q2LZ76 A0A0Q9WSH7 A0A0L0CEC1 A0A146LEQ0 B4QQ98 T1PK69 A0A0A9Y073 A0A1I8MLV5 A0A0A9XYR1 B4HEX3 Q9VTM2 B3NGZ2 A0A1W4W6K0 B4PFM8 A0A0P5DLK9 B4KUS5 E0VAN8 A0A3B0J8Y3 A0A3Q3G744 A0A0P5DFU1 A0A0P5D7I4 A0A2D0SW18 A0A0P5ZRK5 A0A1A8DR56 A0A3B0J9X1
A0A1B0D580 A0A1L8DRK0 A0A1L8DRV4 A0A1J1J1X5 A0A182JIQ6 A0A1B0C9D6 A0A023ESE3 A0A182QRW7 A0A067QWF0 A0A182Y5X8 A0A182U1I3 A0A182SX18 A0A182N9L6 A0A182LUX9 A0A2J7QGS8 A0A182KDX2 A0A2M4ANX9 A0A182LP79 A0A2M4AP09 A0A182WEV6 A0A182RD29 A0A1B6I3C7 A0A2S2R5G8 A0A182FC28 A0A2M3Z7Q1 A0A182VG11 A0A2M3ZI03 A0A2M4BM58 A0A2M4CSS7 A0A2M4CSJ5 A0A182HNC2 A0A1B6FQL0 A0A2M4BM35 A0A182XC23 A0A336K702 A0A2R7VZ90 A0A034VQ42 A0A0K8UBF9 A0A182PL67 A0A0A1WEP6 A0A1Q3F6G1 D6WDC5 J9JPJ3 W8C3A4 A0A069DSX3 A0A0V0GCI1 A0A023FAB1 B0WLD0 B4IZC0 A0A0P4VV67 R4FKY8 A0A1B6D814 E9H287 K7IMS9 A0A0P5ZG97 A0A1A9WH61 B4N4M0 A0A162CC92 A0A0P6A778 A0A0P4YDQ0 A0A1I8P1D5 A0A0P4XZ24 A0A0P5U319 A0A0M3QWZ7 A0A0C9R5G0 B4LCR0 B4HAH3 A0A0P6J0J1 A0A0Q9WKC7 Q2LZ76 A0A0Q9WSH7 A0A0L0CEC1 A0A146LEQ0 B4QQ98 T1PK69 A0A0A9Y073 A0A1I8MLV5 A0A0A9XYR1 B4HEX3 Q9VTM2 B3NGZ2 A0A1W4W6K0 B4PFM8 A0A0P5DLK9 B4KUS5 E0VAN8 A0A3B0J8Y3 A0A3Q3G744 A0A0P5DFU1 A0A0P5D7I4 A0A2D0SW18 A0A0P5ZRK5 A0A1A8DR56 A0A3B0J9X1
PDB
6BXO
E-value=5.62656e-23,
Score=265
Ontologies
PANTHER
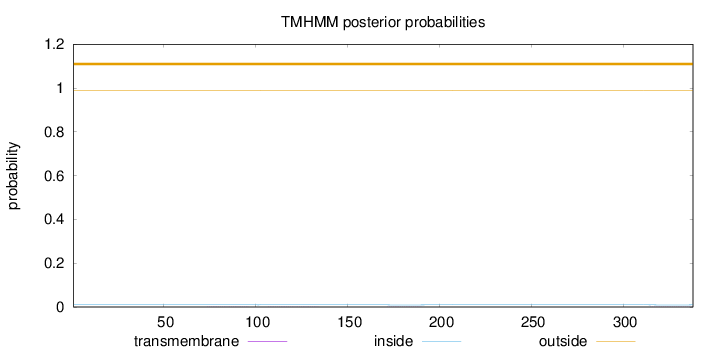
Topology
Length:
338
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03749
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01017
outside
1 - 338
Population Genetic Test Statistics
Pi
333.192769
Theta
236.784723
Tajima's D
1.258885
CLR
0
CSRT
0.727713614319284
Interpretation
Uncertain
