Gene
KWMTBOMO09799
Pre Gene Modal
BGIBMGA013149
Annotation
PREDICTED:_protein_DGCR14_homolog_[Papilio_xuthus]
Full name
Mothers against decapentaplegic homolog
Alternative Name
SMAD family member
Location in the cell
Nuclear Reliability : 4.066
Sequence
CDS
ATGTCAGCTGGAGGAAGCGCTATGAGGCCTGGAGAGGCAGCATTACAGAACATGAAGCTGTTGAAAGCTGAGACTACTGTGTTCAAAGTACCAAAAATACCCAAACAGAAGTCAGAGAAGAAGAGCAAGGTTTTAGAGGAAGATGCTTATGTGGAGGGCATAGCAAAAATTATTCAAAGAGATTTCTTCCCTGACCTAGAGAAACTGAATGCACAGAACGAATACTTGGAGGCTACAGAAAACAAAGACTATCAGAGACTTCGAGAATTAACTCAAAAGTATAGCGGGAACAGACCACCTACAGAACCATATAACACCCCAGCCACTTTTGACACGCCAAACATTGACCGGCCGTGCACTCCTGCTACCAACAATGAGGATGCTCCCAGACAAACAAAAAATGCCACTAAAGACAAAACCGCAGATATTACTGATAACCACAGCTTAGATTCCTTTCTGGCCAAACACACGAGCGAGGACAACGAAAGCTACGATCGAGTTATATCGTTGGAGCAAAAGAAGCGTGCAAATAAAATAGCATCGCAGCTTCAAGCCGAGATCACGTCTGCAACACTCGCAGATGAAGCACTAGCTTTACCCTCCATAGAACAACAAGCTGATCAAACGGAAAGGCCACATGAATTGGACACTTGGCGTTACAAAGCTCGTAATTACATAATGTACGTGCCTGACGGGGCGGAGTCCCAGCGCCCGCCGCCGAAGCCCGAACTAGTCCACCAGAACACAAGGCTAAAAGCTCAACCCTTCGACTACGCTAAGAACAAAGAGACAATAAGCGCCATCGCGAGGAATCAGGACGCGACAGTAAACGGCAGGATAGGCGTGGACGGCAGCAGTTTGAGTTCAGAGAAGGAAGAGCGCTATCCGATACTGCAGACCTCGTCCCCCCGCCCCGGCGAGGGCGCGTCCCCCCTCATGACGTGGGGACACATCGAGGGGACGCCATTCAGACTCGACGGCGGGGACACGCCGCTACCTGCCGCGGGGGCGGGAGTGGCGTACAAGATGCTGGAAGGCGGCGCCCGGGAACGTATCGCGCTGCAGCTCGCCGAGCAACTCGCCAAGAGGAGGCGGCCCACCACCCCGAGGATACCTCCGCCCTCGCCGAGTTTTCGAACGAGCACCGAGCGCCTGGCCAGCATGTCGCCGGCGGCCAGGAAATTAGCCGCGAAACATTTACTAACACCGAGACTCAAGCTGACGCCAAACGACATGGGAATTTTATCGCACGCGGCAACACCGAAACGAACACCAACACGGCGACCGTTAGTGGCAACACCGAGCACGAATAAGAACACGGCAACATCGACCCGTCCTTCCGACTCTGGCAACGTGAACAACAGCAGTACTATGATAGAAAATAATCTCACCGACAACTTGCTGCAGATTAATGTGTGCAAAAGGAATAGATTAAAAGCTAAAGACTTCTTCGAATAA
Protein
MSAGGSAMRPGEAALQNMKLLKAETTVFKVPKIPKQKSEKKSKVLEEDAYVEGIAKIIQRDFFPDLEKLNAQNEYLEATENKDYQRLRELTQKYSGNRPPTEPYNTPATFDTPNIDRPCTPATNNEDAPRQTKNATKDKTADITDNHSLDSFLAKHTSEDNESYDRVISLEQKKRANKIASQLQAEITSATLADEALALPSIEQQADQTERPHELDTWRYKARNYIMYVPDGAESQRPPPKPELVHQNTRLKAQPFDYAKNKETISAIARNQDATVNGRIGVDGSSLSSEKEERYPILQTSSPRPGEGASPLMTWGHIEGTPFRLDGGDTPLPAAGAGVAYKMLEGGARERIALQLAEQLAKRRRPTTPRIPPPSPSFRTSTERLASMSPAARKLAAKHLLTPRLKLTPNDMGILSHAATPKRTPTRRPLVATPSTNKNTATSTRPSDSGNVNNSSTMIENNLTDNLLQINVCKRNRLKAKDFFE
Summary
Similarity
Belongs to the dwarfin/SMAD family.
Uniprot
H9JUD6
A0A2A4K874
A0A194QIL9
A0A3S2NG66
A0A212FMS4
A0A0N1IHV9
+ More
A0A1W4X9M2 A0A182Q9G4 A0A182P2K8 A0A182K0L3 A0A182MES8 N6SXM2 A0A182NSB4 U4UAF3 A0A1B6J8C4 A0A084VC49 A0A067QM37 A0A182GR23 A0A182SKJ5 Q7PXD2 A0A182YGB6 A0A182R4M3 A0A182X9P4 A0A182L8W4 A0A182VBP6 A0A182HKM8 A0A340TBC5 W5J7F1 Q16XN5 A0A0C9QMI0 A0A0L7QL02 A0A182TK09 A0A1B6FM95 A0A0T6BC55 A0A2J7Q6Y0 A0A154PML3 T1GT80 A0A2A3ESM4 A0A182IMD4 A0A182F861 A0A2M4BKZ9 A0A088AUP8 A0A1J1IB59 A0A1B6KB47 A0A2S2QLF9 A0A023F174 A0A026VV06 A0A1B6CZD1 A0A195CS64 A0A158NW43 A0A3L8DV04 A0A151I0U0 A0A151JYC3 A0A034WNX9 A0A195E3Q4 B0WKP9 A0A0K8V7N0 A0A224XMA0 K7IS55 A0A2H8TEC2 F4X2F1 E2BCB6 A0A0L0C2A7 A0A1Q3F4H2 A0A1B0FKV0 A0A0A1X1H8 A0A1B0BWA6 W8B9Z7 A0A1A9XSY2 T1HIG4 A0A151WVF8 A0A0J7L8R9 A0A1A9Z4R9 J9K3J3 A0A1A9VSH3 A0A0P5XLJ8 A0A0P4Z9C6 E2AAH4 A0A0P5ZG05 A0A0P5DAV5 A0A0N8CE25 A0A0P5PFF0 A0A146KW69 A0A0P4Z7P7 A0A0A9YSI1 A0A0P5XER4 E9G8N0 A0A1I8Q4I4 A0A0P5MBT2 A0A1A9WC46 A0A0L8G7L0 A0A139WG28 A0A1I8M1P6 T1J2N2 A0A210PVA4
A0A1W4X9M2 A0A182Q9G4 A0A182P2K8 A0A182K0L3 A0A182MES8 N6SXM2 A0A182NSB4 U4UAF3 A0A1B6J8C4 A0A084VC49 A0A067QM37 A0A182GR23 A0A182SKJ5 Q7PXD2 A0A182YGB6 A0A182R4M3 A0A182X9P4 A0A182L8W4 A0A182VBP6 A0A182HKM8 A0A340TBC5 W5J7F1 Q16XN5 A0A0C9QMI0 A0A0L7QL02 A0A182TK09 A0A1B6FM95 A0A0T6BC55 A0A2J7Q6Y0 A0A154PML3 T1GT80 A0A2A3ESM4 A0A182IMD4 A0A182F861 A0A2M4BKZ9 A0A088AUP8 A0A1J1IB59 A0A1B6KB47 A0A2S2QLF9 A0A023F174 A0A026VV06 A0A1B6CZD1 A0A195CS64 A0A158NW43 A0A3L8DV04 A0A151I0U0 A0A151JYC3 A0A034WNX9 A0A195E3Q4 B0WKP9 A0A0K8V7N0 A0A224XMA0 K7IS55 A0A2H8TEC2 F4X2F1 E2BCB6 A0A0L0C2A7 A0A1Q3F4H2 A0A1B0FKV0 A0A0A1X1H8 A0A1B0BWA6 W8B9Z7 A0A1A9XSY2 T1HIG4 A0A151WVF8 A0A0J7L8R9 A0A1A9Z4R9 J9K3J3 A0A1A9VSH3 A0A0P5XLJ8 A0A0P4Z9C6 E2AAH4 A0A0P5ZG05 A0A0P5DAV5 A0A0N8CE25 A0A0P5PFF0 A0A146KW69 A0A0P4Z7P7 A0A0A9YSI1 A0A0P5XER4 E9G8N0 A0A1I8Q4I4 A0A0P5MBT2 A0A1A9WC46 A0A0L8G7L0 A0A139WG28 A0A1I8M1P6 T1J2N2 A0A210PVA4
Pubmed
19121390
26354079
22118469
23537049
24438588
24845553
+ More
26483478 12364791 14747013 17210077 25244985 20966253 20920257 23761445 17510324 25474469 24508170 21347285 30249741 25348373 20075255 21719571 20798317 26108605 25830018 24495485 26823975 25401762 21292972 18362917 19820115 25315136 28812685
26483478 12364791 14747013 17210077 25244985 20966253 20920257 23761445 17510324 25474469 24508170 21347285 30249741 25348373 20075255 21719571 20798317 26108605 25830018 24495485 26823975 25401762 21292972 18362917 19820115 25315136 28812685
EMBL
BABH01038588
BABH01038589
BABH01038590
NWSH01000035
PCG80427.1
KQ458756
+ More
KPJ05244.1 RSAL01000051 RVE50253.1 AGBW02007651 OWR55023.1 KQ460426 KPJ14813.1 AXCN02000206 AXCM01006934 APGK01052881 KB741216 ENN72509.1 KB631897 ERL86930.1 GECU01012321 JAS95385.1 ATLV01010370 KE524580 KFB35543.1 KK853259 KDR09235.1 JXUM01081707 KQ563264 KXJ74176.1 AAAB01008987 EAA01147.4 APCN01000847 ADMH02002026 ETN59891.1 CH477534 EAT39407.1 GBYB01004714 JAG74481.1 KQ414934 KOC59270.1 GECZ01018441 JAS51328.1 LJIG01002004 KRT84936.1 NEVH01017450 PNF24344.1 KQ434954 KZC12538.1 CAQQ02200218 KZ288192 PBC34216.1 GGFJ01004533 MBW53674.1 CVRI01000047 CRK97527.1 GEBQ01031573 JAT08404.1 GGMS01009361 MBY78564.1 GBBI01003487 JAC15225.1 KK107801 EZA47587.1 GEDC01018482 JAS18816.1 KQ977329 KYN03536.1 ADTU01027791 QOIP01000004 RLU24247.1 KQ976606 KYM79335.1 KQ981477 KYN41453.1 GAKP01003479 JAC55473.1 KQ979701 KYN19504.1 DS231974 EDS29980.1 GDHF01017438 JAI34876.1 GFTR01005468 JAW10958.1 GFXV01000636 MBW12441.1 GL888578 EGI59376.1 GL447257 EFN86652.1 JRES01000984 KNC26493.1 GFDL01012585 JAV22460.1 CCAG010002139 GBXI01009566 JAD04726.1 JXJN01021691 GAMC01011068 JAB95487.1 ACPB03010282 KQ982721 KYQ51661.1 LBMM01000267 KMR02300.1 ABLF02037641 GDIP01196440 GDIP01168296 GDIP01146146 GDIP01088946 GDIP01071906 GDIP01066956 GDIP01065504 JAM31809.1 GDIP01216687 JAJ06715.1 GL438125 EFN69575.1 GDIP01211693 GDIP01185088 GDIP01183734 GDIP01162946 GDIP01137639 GDIP01132293 GDIP01073258 GDIP01044604 JAM59111.1 GDIP01164042 JAJ59360.1 GDIP01213492 GDIP01200534 GDIP01159638 GDIP01146147 GDIP01140847 GDIP01088947 GDIP01039660 JAL62867.1 GDIQ01131544 JAL20182.1 GDHC01017885 JAQ00744.1 GDIP01216685 JAJ06717.1 GBHO01009032 GBHO01009031 GBHO01009029 GBHO01009028 JAG34572.1 JAG34573.1 JAG34575.1 JAG34576.1 GDIP01213493 GDIP01211694 GDIP01185087 GDIP01137685 GDIP01132294 GDIP01073257 GDIP01053720 GDIP01044605 LRGB01002451 JAM30458.1 KZS07545.1 GL732535 EFX84237.1 GDIQ01157769 JAK93956.1 KQ423598 KOF72570.1 KQ971348 KYB26902.1 JH431806 NEDP02005467 OWF40395.1
KPJ05244.1 RSAL01000051 RVE50253.1 AGBW02007651 OWR55023.1 KQ460426 KPJ14813.1 AXCN02000206 AXCM01006934 APGK01052881 KB741216 ENN72509.1 KB631897 ERL86930.1 GECU01012321 JAS95385.1 ATLV01010370 KE524580 KFB35543.1 KK853259 KDR09235.1 JXUM01081707 KQ563264 KXJ74176.1 AAAB01008987 EAA01147.4 APCN01000847 ADMH02002026 ETN59891.1 CH477534 EAT39407.1 GBYB01004714 JAG74481.1 KQ414934 KOC59270.1 GECZ01018441 JAS51328.1 LJIG01002004 KRT84936.1 NEVH01017450 PNF24344.1 KQ434954 KZC12538.1 CAQQ02200218 KZ288192 PBC34216.1 GGFJ01004533 MBW53674.1 CVRI01000047 CRK97527.1 GEBQ01031573 JAT08404.1 GGMS01009361 MBY78564.1 GBBI01003487 JAC15225.1 KK107801 EZA47587.1 GEDC01018482 JAS18816.1 KQ977329 KYN03536.1 ADTU01027791 QOIP01000004 RLU24247.1 KQ976606 KYM79335.1 KQ981477 KYN41453.1 GAKP01003479 JAC55473.1 KQ979701 KYN19504.1 DS231974 EDS29980.1 GDHF01017438 JAI34876.1 GFTR01005468 JAW10958.1 GFXV01000636 MBW12441.1 GL888578 EGI59376.1 GL447257 EFN86652.1 JRES01000984 KNC26493.1 GFDL01012585 JAV22460.1 CCAG010002139 GBXI01009566 JAD04726.1 JXJN01021691 GAMC01011068 JAB95487.1 ACPB03010282 KQ982721 KYQ51661.1 LBMM01000267 KMR02300.1 ABLF02037641 GDIP01196440 GDIP01168296 GDIP01146146 GDIP01088946 GDIP01071906 GDIP01066956 GDIP01065504 JAM31809.1 GDIP01216687 JAJ06715.1 GL438125 EFN69575.1 GDIP01211693 GDIP01185088 GDIP01183734 GDIP01162946 GDIP01137639 GDIP01132293 GDIP01073258 GDIP01044604 JAM59111.1 GDIP01164042 JAJ59360.1 GDIP01213492 GDIP01200534 GDIP01159638 GDIP01146147 GDIP01140847 GDIP01088947 GDIP01039660 JAL62867.1 GDIQ01131544 JAL20182.1 GDHC01017885 JAQ00744.1 GDIP01216685 JAJ06717.1 GBHO01009032 GBHO01009031 GBHO01009029 GBHO01009028 JAG34572.1 JAG34573.1 JAG34575.1 JAG34576.1 GDIP01213493 GDIP01211694 GDIP01185087 GDIP01137685 GDIP01132294 GDIP01073257 GDIP01053720 GDIP01044605 LRGB01002451 JAM30458.1 KZS07545.1 GL732535 EFX84237.1 GDIQ01157769 JAK93956.1 KQ423598 KOF72570.1 KQ971348 KYB26902.1 JH431806 NEDP02005467 OWF40395.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000283053
UP000007151
UP000053240
+ More
UP000192223 UP000075886 UP000075885 UP000075881 UP000075883 UP000019118 UP000075884 UP000030742 UP000030765 UP000027135 UP000069940 UP000249989 UP000075901 UP000007062 UP000076408 UP000075900 UP000076407 UP000075882 UP000075903 UP000075840 UP000075920 UP000000673 UP000008820 UP000053825 UP000075902 UP000235965 UP000076502 UP000015102 UP000242457 UP000075880 UP000069272 UP000005203 UP000183832 UP000053097 UP000078542 UP000005205 UP000279307 UP000078540 UP000078541 UP000078492 UP000002320 UP000002358 UP000007755 UP000008237 UP000037069 UP000092444 UP000092460 UP000092443 UP000015103 UP000075809 UP000036403 UP000092445 UP000007819 UP000078200 UP000000311 UP000076858 UP000000305 UP000095300 UP000091820 UP000053454 UP000007266 UP000095301 UP000242188
UP000192223 UP000075886 UP000075885 UP000075881 UP000075883 UP000019118 UP000075884 UP000030742 UP000030765 UP000027135 UP000069940 UP000249989 UP000075901 UP000007062 UP000076408 UP000075900 UP000076407 UP000075882 UP000075903 UP000075840 UP000075920 UP000000673 UP000008820 UP000053825 UP000075902 UP000235965 UP000076502 UP000015102 UP000242457 UP000075880 UP000069272 UP000005203 UP000183832 UP000053097 UP000078542 UP000005205 UP000279307 UP000078540 UP000078541 UP000078492 UP000002320 UP000002358 UP000007755 UP000008237 UP000037069 UP000092444 UP000092460 UP000092443 UP000015103 UP000075809 UP000036403 UP000092445 UP000007819 UP000078200 UP000000311 UP000076858 UP000000305 UP000095300 UP000091820 UP000053454 UP000007266 UP000095301 UP000242188
PRIDE
Interpro
IPR019148
Nuclear_protein_DGCR14_ESS-2
+ More
IPR008472 DUF753
IPR008967 p53-like_TF_DNA-bd
IPR001699 TF_T-box
IPR018186 TF_T-box_CS
IPR036960 T-box_sf
IPR001132 SMAD_dom_Dwarfin-type
IPR013019 MAD_homology_MH1
IPR017855 SMAD-like_dom_sf
IPR008984 SMAD_FHA_dom_sf
IPR013790 Dwarfin
IPR036578 SMAD_MH1_sf
IPR003619 MAD_homology1_Dwarfin-type
IPR008472 DUF753
IPR008967 p53-like_TF_DNA-bd
IPR001699 TF_T-box
IPR018186 TF_T-box_CS
IPR036960 T-box_sf
IPR001132 SMAD_dom_Dwarfin-type
IPR013019 MAD_homology_MH1
IPR017855 SMAD-like_dom_sf
IPR008984 SMAD_FHA_dom_sf
IPR013790 Dwarfin
IPR036578 SMAD_MH1_sf
IPR003619 MAD_homology1_Dwarfin-type
Gene 3D
CDD
ProteinModelPortal
H9JUD6
A0A2A4K874
A0A194QIL9
A0A3S2NG66
A0A212FMS4
A0A0N1IHV9
+ More
A0A1W4X9M2 A0A182Q9G4 A0A182P2K8 A0A182K0L3 A0A182MES8 N6SXM2 A0A182NSB4 U4UAF3 A0A1B6J8C4 A0A084VC49 A0A067QM37 A0A182GR23 A0A182SKJ5 Q7PXD2 A0A182YGB6 A0A182R4M3 A0A182X9P4 A0A182L8W4 A0A182VBP6 A0A182HKM8 A0A340TBC5 W5J7F1 Q16XN5 A0A0C9QMI0 A0A0L7QL02 A0A182TK09 A0A1B6FM95 A0A0T6BC55 A0A2J7Q6Y0 A0A154PML3 T1GT80 A0A2A3ESM4 A0A182IMD4 A0A182F861 A0A2M4BKZ9 A0A088AUP8 A0A1J1IB59 A0A1B6KB47 A0A2S2QLF9 A0A023F174 A0A026VV06 A0A1B6CZD1 A0A195CS64 A0A158NW43 A0A3L8DV04 A0A151I0U0 A0A151JYC3 A0A034WNX9 A0A195E3Q4 B0WKP9 A0A0K8V7N0 A0A224XMA0 K7IS55 A0A2H8TEC2 F4X2F1 E2BCB6 A0A0L0C2A7 A0A1Q3F4H2 A0A1B0FKV0 A0A0A1X1H8 A0A1B0BWA6 W8B9Z7 A0A1A9XSY2 T1HIG4 A0A151WVF8 A0A0J7L8R9 A0A1A9Z4R9 J9K3J3 A0A1A9VSH3 A0A0P5XLJ8 A0A0P4Z9C6 E2AAH4 A0A0P5ZG05 A0A0P5DAV5 A0A0N8CE25 A0A0P5PFF0 A0A146KW69 A0A0P4Z7P7 A0A0A9YSI1 A0A0P5XER4 E9G8N0 A0A1I8Q4I4 A0A0P5MBT2 A0A1A9WC46 A0A0L8G7L0 A0A139WG28 A0A1I8M1P6 T1J2N2 A0A210PVA4
A0A1W4X9M2 A0A182Q9G4 A0A182P2K8 A0A182K0L3 A0A182MES8 N6SXM2 A0A182NSB4 U4UAF3 A0A1B6J8C4 A0A084VC49 A0A067QM37 A0A182GR23 A0A182SKJ5 Q7PXD2 A0A182YGB6 A0A182R4M3 A0A182X9P4 A0A182L8W4 A0A182VBP6 A0A182HKM8 A0A340TBC5 W5J7F1 Q16XN5 A0A0C9QMI0 A0A0L7QL02 A0A182TK09 A0A1B6FM95 A0A0T6BC55 A0A2J7Q6Y0 A0A154PML3 T1GT80 A0A2A3ESM4 A0A182IMD4 A0A182F861 A0A2M4BKZ9 A0A088AUP8 A0A1J1IB59 A0A1B6KB47 A0A2S2QLF9 A0A023F174 A0A026VV06 A0A1B6CZD1 A0A195CS64 A0A158NW43 A0A3L8DV04 A0A151I0U0 A0A151JYC3 A0A034WNX9 A0A195E3Q4 B0WKP9 A0A0K8V7N0 A0A224XMA0 K7IS55 A0A2H8TEC2 F4X2F1 E2BCB6 A0A0L0C2A7 A0A1Q3F4H2 A0A1B0FKV0 A0A0A1X1H8 A0A1B0BWA6 W8B9Z7 A0A1A9XSY2 T1HIG4 A0A151WVF8 A0A0J7L8R9 A0A1A9Z4R9 J9K3J3 A0A1A9VSH3 A0A0P5XLJ8 A0A0P4Z9C6 E2AAH4 A0A0P5ZG05 A0A0P5DAV5 A0A0N8CE25 A0A0P5PFF0 A0A146KW69 A0A0P4Z7P7 A0A0A9YSI1 A0A0P5XER4 E9G8N0 A0A1I8Q4I4 A0A0P5MBT2 A0A1A9WC46 A0A0L8G7L0 A0A139WG28 A0A1I8M1P6 T1J2N2 A0A210PVA4
Ontologies
Topology
Subcellular location
Nucleus
Cytoplasm
Cytoplasm
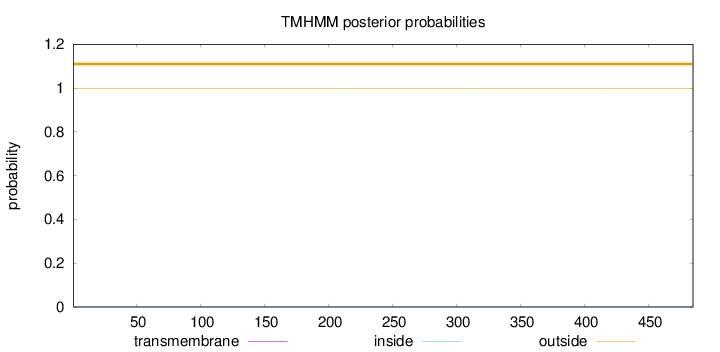
Length:
485
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00159
outside
1 - 485
Population Genetic Test Statistics
Pi
248.048222
Theta
164.444994
Tajima's D
1.030524
CLR
1.686265
CSRT
0.660366981650917
Interpretation
Uncertain
