Gene
KWMTBOMO09798
Pre Gene Modal
BGIBMGA013113
Annotation
PREDICTED:_uncharacterized_protein_LOC106135229_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 2.974
Sequence
CDS
ATGGAGCTACCGGAATGGTTCGACGAAAAACAATTTAACCAGGCGCGGAAGTTCTACGACGAGAACAGCTATTCGTTCTCGTCGTCCATGCTACAGGGTCTAGTGGCTGTTCTCTCGGTGCCTTCTATACTCCGTGTCCTGGTCGGCACGCGACGCTCCAGCTCCGTTTTCACAGCCTACAAACGGTATCTGTCCACGCTTCTGCACACGATCTCCTGGTTCGAACACGAACTGAAGCCCGGAAGCATGTCCTGGAGGTCACTGTACGCAGTGAGGAGTCGCCACGTCAAAGCATCATTAGCCTCAAACGCTAAAGGAACCGGAGTGGTATCGCAGAGAGACATCGCTCTCACGCAATTCGGCTTCATCGGATTCTCGGTACTAAAACCCGACTTCTTTGGCATAAGGCAGCTGGGCGACGGCGACTGGCAAGCTTACAATCACTTCTGGAGGGTCATAGGATACATGATAGGACTGGATGACAGGTACAACATTTGCCGTGAAACAATAGAGGAGACTCGCGAAGTATGCCAATTGCTATTGGACCGAGTTTACACGCCGTGTCTGGAGAATGCGCCCGAGTATTTCGAGCATATGGCCCGCGTCATGCTCGACGGTATGTGGTGCATCAACCCTACAGTCGACCTCGAAGCCAACATGTACGGATGCCGATACTTGGCCGACGTCCCGGGGTACATCTACACGGAGCGCGACAGGATAGAGCTACAACAGAAATTGAGGAAGCATTTAAAAGGGAAATCCCTTGATACCGGCGTAGACTCGTCGCTGCTGATGAGCCAACCAGCCGTCGATGGTCTCCCGGAACGGGCGCCGCGTCTCCTCTACTACAACGACTACAACACGATCGAGACCGCACCGGCCTACAAACAGCTCGGCCTCAAATCGAAATATAAACTGGCGTGCTTCAACATATACATGTGCCTCTATACCACGTTCATTGGTCGTTGGTATTTCAACGCCAATTTCAAATTCAGCTTGTTCCTAATGAGATACTTCCCGTTTTTGGCGTTCTTCAGATTCGGCGTTAGGAATTCATACGTGAATATTTTCGTGGAGGACCTTACATCGGACACTAAGCCTAAGATCAACTCTGAATATTATAAGCCACAGGAGCCGACGCCGTGGTACAGAACAGTTTTGGAACTGCTATTTTAA
Protein
MELPEWFDEKQFNQARKFYDENSYSFSSSMLQGLVAVLSVPSILRVLVGTRRSSSVFTAYKRYLSTLLHTISWFEHELKPGSMSWRSLYAVRSRHVKASLASNAKGTGVVSQRDIALTQFGFIGFSVLKPDFFGIRQLGDGDWQAYNHFWRVIGYMIGLDDRYNICRETIEETREVCQLLLDRVYTPCLENAPEYFEHMARVMLDGMWCINPTVDLEANMYGCRYLADVPGYIYTERDRIELQQKLRKHLKGKSLDTGVDSSLLMSQPAVDGLPERAPRLLYYNDYNTIETAPAYKQLGLKSKYKLACFNIYMCLYTTFIGRWYFNANFKFSLFLMRYFPFLAFFRFGVRNSYVNIFVEDLTSDTKPKINSEYYKPQEPTPWYRTVLELLF
Summary
Uniprot
H9JUA1
A0A212FMP3
A0A437BII2
A0A0N1PII0
A0A2H1WQC7
A0A194REG8
+ More
A0A194QK08 A0A2A4K8T5 A0A194QIA3 A0A194REU2 A0A194QK14 A0A0N0PBN0 A0A212FMV3 A0A194PJJ0 A0A0N1PGM9 A0A194QP37 A0A194QIM4 A0A0L7K374 B0WPZ3 U5ETF9 A0A1L8D9S8 A0A1Q3FAE0 A0A1Q3FB47 A0A182JLM2 A0A1B0GK02 W5J8F7 A0A2M4ATA7 A0A084W2A5 B0WSR0 A0A1Y9H265 A0A182R3F6 A0A182QLL0 A0A182F9M7 A0A182WAG4 A0A1J1HRI2 A0A182S5H9 A0A1J1HQ27 A0A182YIF8 A0A182G7D7 A0A182PCJ1 A0A0K8VM12 W8AVQ4 A0A034VU34 W8B661 Q7Q9K9 A0A182HP11 A0A182VGI6 A0A182LBJ8 B0WPZ1 B0WPZ2 A0A3B0JNX8 B4LY28 A0A3B0KE42 Q16KG8 A0A182XEY6 A0A023EPG7 Q294C6 A0A182JV93 B4GL98 A0A0M3QYJ7 A0A182M5B7 A0A182GGP8 B3P0S0 T1PDT0 B4PS77 A0A0L0C864 B4KAG7 A0A1I8MYZ7 B4QZ39 Q95U32 B4JGB4 A0A1W4W0I1 A0A1I8PFG1 A0A1A9ZR16 A0A1A9UF29 B4NK77 B4HE06 B3MX99 A0A1B0ET71 A0A182G7D8 A0A2S2QIK7
A0A194QK08 A0A2A4K8T5 A0A194QIA3 A0A194REU2 A0A194QK14 A0A0N0PBN0 A0A212FMV3 A0A194PJJ0 A0A0N1PGM9 A0A194QP37 A0A194QIM4 A0A0L7K374 B0WPZ3 U5ETF9 A0A1L8D9S8 A0A1Q3FAE0 A0A1Q3FB47 A0A182JLM2 A0A1B0GK02 W5J8F7 A0A2M4ATA7 A0A084W2A5 B0WSR0 A0A1Y9H265 A0A182R3F6 A0A182QLL0 A0A182F9M7 A0A182WAG4 A0A1J1HRI2 A0A182S5H9 A0A1J1HQ27 A0A182YIF8 A0A182G7D7 A0A182PCJ1 A0A0K8VM12 W8AVQ4 A0A034VU34 W8B661 Q7Q9K9 A0A182HP11 A0A182VGI6 A0A182LBJ8 B0WPZ1 B0WPZ2 A0A3B0JNX8 B4LY28 A0A3B0KE42 Q16KG8 A0A182XEY6 A0A023EPG7 Q294C6 A0A182JV93 B4GL98 A0A0M3QYJ7 A0A182M5B7 A0A182GGP8 B3P0S0 T1PDT0 B4PS77 A0A0L0C864 B4KAG7 A0A1I8MYZ7 B4QZ39 Q95U32 B4JGB4 A0A1W4W0I1 A0A1I8PFG1 A0A1A9ZR16 A0A1A9UF29 B4NK77 B4HE06 B3MX99 A0A1B0ET71 A0A182G7D8 A0A2S2QIK7
Pubmed
EMBL
BABH01038591
BABH01038592
AGBW02007651
OWR55022.1
RSAL01000051
RVE50252.1
+ More
LADJ01000510 KPJ20836.1 ODYU01010247 SOQ55228.1 KQ460323 KPJ15982.1 KQ458756 KPJ05245.1 NWSH01000035 PCG80416.1 KPJ05247.1 KPJ15979.1 KPJ05250.1 KQ460942 KPJ10572.1 OWR55020.1 KQ459602 KPI93203.1 KQ461178 KPJ07758.1 KPJ05246.1 KPJ05249.1 JTDY01012591 KOB52250.1 DS232031 EDS32583.1 GANO01004391 JAB55480.1 GFDF01010851 JAV03233.1 GFDL01010556 JAV24489.1 GFDL01010265 JAV24780.1 AJWK01022481 ADMH02001867 ETN60742.1 GGFK01010700 MBW44021.1 ATLV01019571 KE525275 KFB44349.1 DS232075 EDS34019.1 AXCN02000996 CVRI01000017 CRK90138.1 CRK90141.1 JXUM01046460 JXUM01046461 KQ561478 KXJ78460.1 GDHF01030652 GDHF01012392 JAI21662.1 JAI39922.1 GAMC01017752 JAB88803.1 GAKP01012988 GAKP01012986 JAC45964.1 GAMC01017754 JAB88801.1 AAAB01008900 EAA09433.3 APCN01002864 EDS32581.1 EDS32582.1 OUUW01000008 SPP83947.1 CH940650 EDW66894.2 SPP83946.1 CH477959 EAT34785.1 GAPW01002216 JAC11382.1 CM000070 EAL29038.3 CH479185 EDW38322.1 CP012526 ALC47739.1 AXCM01001086 JXUM01062239 JXUM01062240 KQ562190 KXJ76458.1 CH954181 EDV48968.1 KA646932 AFP61561.1 CM000160 EDW97497.1 JRES01000776 KNC28422.1 CH933806 EDW15680.1 CM000364 EDX12867.1 AY058343 AE014297 AAL13572.1 AAN13634.1 CH916369 EDV92583.1 CH964272 EDW85119.1 CH480815 EDW42098.1 CH902629 EDV35322.1 CCAG010021199 CCAG010021200 CCAG010021201 JXUM01046462 KXJ78461.1 GGMS01008373 MBY77576.1
LADJ01000510 KPJ20836.1 ODYU01010247 SOQ55228.1 KQ460323 KPJ15982.1 KQ458756 KPJ05245.1 NWSH01000035 PCG80416.1 KPJ05247.1 KPJ15979.1 KPJ05250.1 KQ460942 KPJ10572.1 OWR55020.1 KQ459602 KPI93203.1 KQ461178 KPJ07758.1 KPJ05246.1 KPJ05249.1 JTDY01012591 KOB52250.1 DS232031 EDS32583.1 GANO01004391 JAB55480.1 GFDF01010851 JAV03233.1 GFDL01010556 JAV24489.1 GFDL01010265 JAV24780.1 AJWK01022481 ADMH02001867 ETN60742.1 GGFK01010700 MBW44021.1 ATLV01019571 KE525275 KFB44349.1 DS232075 EDS34019.1 AXCN02000996 CVRI01000017 CRK90138.1 CRK90141.1 JXUM01046460 JXUM01046461 KQ561478 KXJ78460.1 GDHF01030652 GDHF01012392 JAI21662.1 JAI39922.1 GAMC01017752 JAB88803.1 GAKP01012988 GAKP01012986 JAC45964.1 GAMC01017754 JAB88801.1 AAAB01008900 EAA09433.3 APCN01002864 EDS32581.1 EDS32582.1 OUUW01000008 SPP83947.1 CH940650 EDW66894.2 SPP83946.1 CH477959 EAT34785.1 GAPW01002216 JAC11382.1 CM000070 EAL29038.3 CH479185 EDW38322.1 CP012526 ALC47739.1 AXCM01001086 JXUM01062239 JXUM01062240 KQ562190 KXJ76458.1 CH954181 EDV48968.1 KA646932 AFP61561.1 CM000160 EDW97497.1 JRES01000776 KNC28422.1 CH933806 EDW15680.1 CM000364 EDX12867.1 AY058343 AE014297 AAL13572.1 AAN13634.1 CH916369 EDV92583.1 CH964272 EDW85119.1 CH480815 EDW42098.1 CH902629 EDV35322.1 CCAG010021199 CCAG010021200 CCAG010021201 JXUM01046462 KXJ78461.1 GGMS01008373 MBY77576.1
Proteomes
UP000005204
UP000007151
UP000283053
UP000053240
UP000053268
UP000218220
+ More
UP000037510 UP000002320 UP000075880 UP000092461 UP000000673 UP000030765 UP000075884 UP000075900 UP000075886 UP000069272 UP000075920 UP000183832 UP000075901 UP000076408 UP000069940 UP000249989 UP000075885 UP000007062 UP000075840 UP000075903 UP000075882 UP000268350 UP000008792 UP000008820 UP000076407 UP000001819 UP000075881 UP000008744 UP000092553 UP000075883 UP000008711 UP000002282 UP000037069 UP000009192 UP000095301 UP000000304 UP000000803 UP000001070 UP000192221 UP000095300 UP000092445 UP000078200 UP000007798 UP000001292 UP000007801 UP000092444
UP000037510 UP000002320 UP000075880 UP000092461 UP000000673 UP000030765 UP000075884 UP000075900 UP000075886 UP000069272 UP000075920 UP000183832 UP000075901 UP000076408 UP000069940 UP000249989 UP000075885 UP000007062 UP000075840 UP000075903 UP000075882 UP000268350 UP000008792 UP000008820 UP000076407 UP000001819 UP000075881 UP000008744 UP000092553 UP000075883 UP000008711 UP000002282 UP000037069 UP000009192 UP000095301 UP000000304 UP000000803 UP000001070 UP000192221 UP000095300 UP000092445 UP000078200 UP000007798 UP000001292 UP000007801 UP000092444
Interpro
Gene 3D
ProteinModelPortal
H9JUA1
A0A212FMP3
A0A437BII2
A0A0N1PII0
A0A2H1WQC7
A0A194REG8
+ More
A0A194QK08 A0A2A4K8T5 A0A194QIA3 A0A194REU2 A0A194QK14 A0A0N0PBN0 A0A212FMV3 A0A194PJJ0 A0A0N1PGM9 A0A194QP37 A0A194QIM4 A0A0L7K374 B0WPZ3 U5ETF9 A0A1L8D9S8 A0A1Q3FAE0 A0A1Q3FB47 A0A182JLM2 A0A1B0GK02 W5J8F7 A0A2M4ATA7 A0A084W2A5 B0WSR0 A0A1Y9H265 A0A182R3F6 A0A182QLL0 A0A182F9M7 A0A182WAG4 A0A1J1HRI2 A0A182S5H9 A0A1J1HQ27 A0A182YIF8 A0A182G7D7 A0A182PCJ1 A0A0K8VM12 W8AVQ4 A0A034VU34 W8B661 Q7Q9K9 A0A182HP11 A0A182VGI6 A0A182LBJ8 B0WPZ1 B0WPZ2 A0A3B0JNX8 B4LY28 A0A3B0KE42 Q16KG8 A0A182XEY6 A0A023EPG7 Q294C6 A0A182JV93 B4GL98 A0A0M3QYJ7 A0A182M5B7 A0A182GGP8 B3P0S0 T1PDT0 B4PS77 A0A0L0C864 B4KAG7 A0A1I8MYZ7 B4QZ39 Q95U32 B4JGB4 A0A1W4W0I1 A0A1I8PFG1 A0A1A9ZR16 A0A1A9UF29 B4NK77 B4HE06 B3MX99 A0A1B0ET71 A0A182G7D8 A0A2S2QIK7
A0A194QK08 A0A2A4K8T5 A0A194QIA3 A0A194REU2 A0A194QK14 A0A0N0PBN0 A0A212FMV3 A0A194PJJ0 A0A0N1PGM9 A0A194QP37 A0A194QIM4 A0A0L7K374 B0WPZ3 U5ETF9 A0A1L8D9S8 A0A1Q3FAE0 A0A1Q3FB47 A0A182JLM2 A0A1B0GK02 W5J8F7 A0A2M4ATA7 A0A084W2A5 B0WSR0 A0A1Y9H265 A0A182R3F6 A0A182QLL0 A0A182F9M7 A0A182WAG4 A0A1J1HRI2 A0A182S5H9 A0A1J1HQ27 A0A182YIF8 A0A182G7D7 A0A182PCJ1 A0A0K8VM12 W8AVQ4 A0A034VU34 W8B661 Q7Q9K9 A0A182HP11 A0A182VGI6 A0A182LBJ8 B0WPZ1 B0WPZ2 A0A3B0JNX8 B4LY28 A0A3B0KE42 Q16KG8 A0A182XEY6 A0A023EPG7 Q294C6 A0A182JV93 B4GL98 A0A0M3QYJ7 A0A182M5B7 A0A182GGP8 B3P0S0 T1PDT0 B4PS77 A0A0L0C864 B4KAG7 A0A1I8MYZ7 B4QZ39 Q95U32 B4JGB4 A0A1W4W0I1 A0A1I8PFG1 A0A1A9ZR16 A0A1A9UF29 B4NK77 B4HE06 B3MX99 A0A1B0ET71 A0A182G7D8 A0A2S2QIK7
PDB
5O1M
E-value=0.000312029,
Score=104
Ontologies
PANTHER
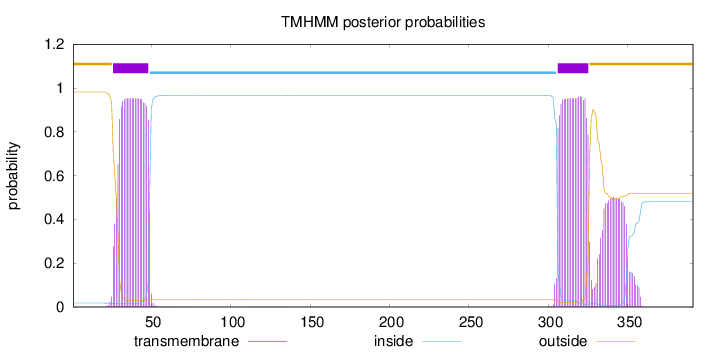
Topology
Length:
391
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
49.20497
Exp number, first 60 AAs:
20.18563
Total prob of N-in:
0.01808
POSSIBLE N-term signal
sequence
outside
1 - 25
TMhelix
26 - 48
inside
49 - 305
TMhelix
306 - 325
outside
326 - 391
Population Genetic Test Statistics
Pi
288.445044
Theta
166.53497
Tajima's D
2.295371
CLR
0.171484
CSRT
0.921953902304885
Interpretation
Uncertain
