Gene
KWMTBOMO09793
Pre Gene Modal
BGIBMGA013110
Annotation
PREDICTED:_SHC_SH2_domain-binding_protein_1_isoform_X2_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.761 Nuclear Reliability : 1.083
Sequence
CDS
ATGCCTTCCGTCTATACATTTTCCCGGAGTGATAACGAGATACTTCAAGAGTTGCTGAAGGTATTTAACACGGCAAGGGGCACAGCACGAGAACAGTGGTCCATACAGGCCGAGCTGTTAGTGGAACCAGTTGGATGGGATGCGCTTTGGAAATTATCAAAAGAATTTTGTAAGAAATTCGAAGTTCGCTACCCTTGCATAGCATATGTCACAGTCACTTCAGTGGACTTTGAGAACTTGTCGTCATCTGTCGATGTACTAAGCGTGCAGCATGAAGCAGTCACTCTCCCGGAGTCAGTTGTCGATGTTCCACTAGTGGAATTGTGGCCAACCACAAAACAACGTGAACAATGTGTGAATGCTGCAACCACTGCAGAGTTCATTGATCTACTCAGATTTTATTACAACGACATTTGGATGCCATGGGATGATCAGGATGATAAGATCCTATTACCGAACACGGTAGAAGACCGTATGCAGCTCTGGTCTGACATGCACAATGGGACCATACAACACTGTGTAGCACGATCTATAACATTACTGAGAAGTAGTGCAATAGATGCACACAATAAACTAAATGTTTTAGATTCTTCACTGTGTGAAGAGGATGTGGAAAATGATGATGATTCACTTCTACCCCCGAACTACATATCACAATGTGCAGAGTTAAATGCTCGTTTAGACGGGCTTATGTCGCAGTGGACACTGTATGAAAACCCTCTGATCAGAGAACAGTATTTGGCCAAAATGAAACACAAATGGCAAAAAAAGAAAAGTAAGAAAAATGTGGTAGCTCTCTGGCAAGGAGGTTCAATATTTGAATTCGACGAGATCTCTAAATTCCTTCGTTCTCGTCTCACCAATGAATGCACCTTGACCGTAATGGTCTCGGCAGAAGACGGCCTCACTCTCGAACCGGAGGAAGTCGTGGTGTGCAGCAAACATTACGAAATACCAGAAATGCCTCTTCTTCAAATATCTATATGCAGTTATAAAGGCGCGACCCTAAGGGCGACCGACATGCGTTCGTGGCTGCTGATGCTAAGTGAGGACTGTCGTCTGCGCGACCTGACCCTGCACTGCGCCCTAGTCAACACCGTGCTGCTGATGCGAGCCGGGACGTTGCATATAACCAATTGCGCCTTCCTGGATGATTCAAAAAATGCACAGAGTGATTTTGCGCAAGGCATAGTTGCCATGTCGGGTGCTAAGATCCTGATCGAAGACTGTACATTTGATAATTTCTATTCCGGCATAGTCGTACATAATGGGGCTCAGGTGGAGCTTCGTAACTGCATCATCAAACGTTGCGGCGTCGGGATCCAGATGTATTGGGGCGCGCGCGTCGAGCTGAGCGCCACCACGATCATGGACTGCGCCGAGAACAGCATACGTTGCGAGGTGGAAGCCGGCGCTTGTTCGAAATACACTGATGTGAAAGGTCTACAAATTAAGACGAATTGCAGAATCGGTAGCGGAAACCTTTCGAAGGAGGTACTTGTCGTCGGACAAGAAGCGACGATCATTTGA
Protein
MPSVYTFSRSDNEILQELLKVFNTARGTAREQWSIQAELLVEPVGWDALWKLSKEFCKKFEVRYPCIAYVTVTSVDFENLSSSVDVLSVQHEAVTLPESVVDVPLVELWPTTKQREQCVNAATTAEFIDLLRFYYNDIWMPWDDQDDKILLPNTVEDRMQLWSDMHNGTIQHCVARSITLLRSSAIDAHNKLNVLDSSLCEEDVENDDDSLLPPNYISQCAELNARLDGLMSQWTLYENPLIREQYLAKMKHKWQKKKSKKNVVALWQGGSIFEFDEISKFLRSRLTNECTLTVMVSAEDGLTLEPEEVVVCSKHYEIPEMPLLQISICSYKGATLRATDMRSWLLMLSEDCRLRDLTLHCALVNTVLLMRAGTLHITNCAFLDDSKNAQSDFAQGIVAMSGAKILIEDCTFDNFYSGIVVHNGAQVELRNCIIKRCGVGIQMYWGARVELSATTIMDCAENSIRCEVEAGACSKYTDVKGLQIKTNCRIGSGNLSKEVLVVGQEATII
Summary
Uniprot
A0A2A4JW47
A0A3S2NAD1
A0A212FBQ1
A0A2W1B8K1
A0A194Q1D7
S4PAE8
+ More
A0A0N0PAP9 A0A2H1VH86 A0A2J7QBL5 K7IM45 A0A232FG39 A0A1J1HS36 A0A088A1B9 A0A067QNE5 A0A0L7R5P8 A0A0M9A026 A0A0C9QWR0 A0A310SJF3 A0A2A3EM40 A0A1A9WEX0 A0A151I693 A0A154P6V9 A0A195FWM9 A0A195E3S2 A0A026WVV6 A0A151XA95 A0A158NNB7 F4WLG8 D6WKG0 A0A195AUI8 N6U0L2 A0A0J7P2B6 U4TY03 A0A2J7QBL1 A0A182JM71 A0A182HFI6 E9IUG5 A0A0K8W6D6 A0A182MRN2 A0A182P4V8 A0A1Y1M6F4 A0A084VND7 A0A0T6AVE5 A0A182F615 Q17JG4 A0A182W7A4 A0A182Q578 A0A182GSK8 E1ZZV0 A0A182RC82 A0A1W4WNU1 A0A182WU99 A0A182L1Q6 A0A182HUW7 B0W7M5 A0A2P8Z2J0 A0A182USE2 A0A1S4HDJ6 A0A182TIB1 A0A1B6FCS6 A0A1Q3EXJ2 A0A1B6KS84 E2C3T7 A0A1B6DZD4 A0A0A1XA85 A0A1Y1M2V5 A0A182Y021 A0A336LQZ3
A0A0N0PAP9 A0A2H1VH86 A0A2J7QBL5 K7IM45 A0A232FG39 A0A1J1HS36 A0A088A1B9 A0A067QNE5 A0A0L7R5P8 A0A0M9A026 A0A0C9QWR0 A0A310SJF3 A0A2A3EM40 A0A1A9WEX0 A0A151I693 A0A154P6V9 A0A195FWM9 A0A195E3S2 A0A026WVV6 A0A151XA95 A0A158NNB7 F4WLG8 D6WKG0 A0A195AUI8 N6U0L2 A0A0J7P2B6 U4TY03 A0A2J7QBL1 A0A182JM71 A0A182HFI6 E9IUG5 A0A0K8W6D6 A0A182MRN2 A0A182P4V8 A0A1Y1M6F4 A0A084VND7 A0A0T6AVE5 A0A182F615 Q17JG4 A0A182W7A4 A0A182Q578 A0A182GSK8 E1ZZV0 A0A182RC82 A0A1W4WNU1 A0A182WU99 A0A182L1Q6 A0A182HUW7 B0W7M5 A0A2P8Z2J0 A0A182USE2 A0A1S4HDJ6 A0A182TIB1 A0A1B6FCS6 A0A1Q3EXJ2 A0A1B6KS84 E2C3T7 A0A1B6DZD4 A0A0A1XA85 A0A1Y1M2V5 A0A182Y021 A0A336LQZ3
Pubmed
EMBL
NWSH01000564
PCG75620.1
RSAL01000342
RVE42387.1
AGBW02009293
OWR51171.1
+ More
KZ150230 PZC72012.1 KQ459582 KPI98804.1 GAIX01005056 JAA87504.1 KQ461183 KPJ07546.1 ODYU01002515 SOQ40111.1 NEVH01016296 PNF25972.1 NNAY01000259 OXU29662.1 CVRI01000020 CRK90829.1 KK853129 KDR10984.1 KQ414652 KOC66139.1 KQ435803 KOX73133.1 GBYB01008169 GBYB01011596 JAG77936.1 JAG81363.1 KQ764881 OAD54316.1 KZ288215 PBC32767.1 KQ978495 KYM93637.1 KQ434819 KZC06928.1 KQ981208 KYN44722.1 KQ979685 KYN19825.1 KK107109 EZA59229.1 KQ982351 KYQ57295.1 ADTU01021203 GL888208 EGI64904.1 KQ971342 EFA03585.1 KQ976738 KYM75846.1 APGK01054197 KB741247 ENN72067.1 LBMM01000319 KMR01387.1 KB631740 ERL85697.1 PNF25973.1 JXUM01134344 KQ568107 KXJ69138.1 GL765974 EFZ15786.1 GDHF01005677 JAI46637.1 AXCM01001558 GEZM01039091 JAV81419.1 ATLV01014737 KE524984 KFB39481.1 LJIG01022701 KRT79168.1 CH477232 EAT46876.1 AXCN02000801 JXUM01017001 KQ560472 KXJ82232.1 GL435529 EFN73290.1 APCN01002480 DS231854 EDS38060.1 PYGN01000224 PSN50703.1 AAAB01008964 GECZ01021750 JAS48019.1 GFDL01015127 JAV19918.1 GEBQ01025689 JAT14288.1 GL452364 EFN77323.1 GEDC01006301 JAS30997.1 GBXI01006647 JAD07645.1 GEZM01042507 JAV79811.1 UFQS01000114 UFQT01000114 SSW99855.1 SSX20235.1
KZ150230 PZC72012.1 KQ459582 KPI98804.1 GAIX01005056 JAA87504.1 KQ461183 KPJ07546.1 ODYU01002515 SOQ40111.1 NEVH01016296 PNF25972.1 NNAY01000259 OXU29662.1 CVRI01000020 CRK90829.1 KK853129 KDR10984.1 KQ414652 KOC66139.1 KQ435803 KOX73133.1 GBYB01008169 GBYB01011596 JAG77936.1 JAG81363.1 KQ764881 OAD54316.1 KZ288215 PBC32767.1 KQ978495 KYM93637.1 KQ434819 KZC06928.1 KQ981208 KYN44722.1 KQ979685 KYN19825.1 KK107109 EZA59229.1 KQ982351 KYQ57295.1 ADTU01021203 GL888208 EGI64904.1 KQ971342 EFA03585.1 KQ976738 KYM75846.1 APGK01054197 KB741247 ENN72067.1 LBMM01000319 KMR01387.1 KB631740 ERL85697.1 PNF25973.1 JXUM01134344 KQ568107 KXJ69138.1 GL765974 EFZ15786.1 GDHF01005677 JAI46637.1 AXCM01001558 GEZM01039091 JAV81419.1 ATLV01014737 KE524984 KFB39481.1 LJIG01022701 KRT79168.1 CH477232 EAT46876.1 AXCN02000801 JXUM01017001 KQ560472 KXJ82232.1 GL435529 EFN73290.1 APCN01002480 DS231854 EDS38060.1 PYGN01000224 PSN50703.1 AAAB01008964 GECZ01021750 JAS48019.1 GFDL01015127 JAV19918.1 GEBQ01025689 JAT14288.1 GL452364 EFN77323.1 GEDC01006301 JAS30997.1 GBXI01006647 JAD07645.1 GEZM01042507 JAV79811.1 UFQS01000114 UFQT01000114 SSW99855.1 SSX20235.1
Proteomes
UP000218220
UP000283053
UP000007151
UP000053268
UP000053240
UP000235965
+ More
UP000002358 UP000215335 UP000183832 UP000005203 UP000027135 UP000053825 UP000053105 UP000242457 UP000091820 UP000078542 UP000076502 UP000078541 UP000078492 UP000053097 UP000075809 UP000005205 UP000007755 UP000007266 UP000078540 UP000019118 UP000036403 UP000030742 UP000075880 UP000069940 UP000249989 UP000075883 UP000075885 UP000030765 UP000069272 UP000008820 UP000075920 UP000075886 UP000000311 UP000075900 UP000192223 UP000076407 UP000075882 UP000075840 UP000002320 UP000245037 UP000075903 UP000075902 UP000008237 UP000076408
UP000002358 UP000215335 UP000183832 UP000005203 UP000027135 UP000053825 UP000053105 UP000242457 UP000091820 UP000078542 UP000076502 UP000078541 UP000078492 UP000053097 UP000075809 UP000005205 UP000007755 UP000007266 UP000078540 UP000019118 UP000036403 UP000030742 UP000075880 UP000069940 UP000249989 UP000075883 UP000075885 UP000030765 UP000069272 UP000008820 UP000075920 UP000075886 UP000000311 UP000075900 UP000192223 UP000076407 UP000075882 UP000075840 UP000002320 UP000245037 UP000075903 UP000075902 UP000008237 UP000076408
Pfam
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2A4JW47
A0A3S2NAD1
A0A212FBQ1
A0A2W1B8K1
A0A194Q1D7
S4PAE8
+ More
A0A0N0PAP9 A0A2H1VH86 A0A2J7QBL5 K7IM45 A0A232FG39 A0A1J1HS36 A0A088A1B9 A0A067QNE5 A0A0L7R5P8 A0A0M9A026 A0A0C9QWR0 A0A310SJF3 A0A2A3EM40 A0A1A9WEX0 A0A151I693 A0A154P6V9 A0A195FWM9 A0A195E3S2 A0A026WVV6 A0A151XA95 A0A158NNB7 F4WLG8 D6WKG0 A0A195AUI8 N6U0L2 A0A0J7P2B6 U4TY03 A0A2J7QBL1 A0A182JM71 A0A182HFI6 E9IUG5 A0A0K8W6D6 A0A182MRN2 A0A182P4V8 A0A1Y1M6F4 A0A084VND7 A0A0T6AVE5 A0A182F615 Q17JG4 A0A182W7A4 A0A182Q578 A0A182GSK8 E1ZZV0 A0A182RC82 A0A1W4WNU1 A0A182WU99 A0A182L1Q6 A0A182HUW7 B0W7M5 A0A2P8Z2J0 A0A182USE2 A0A1S4HDJ6 A0A182TIB1 A0A1B6FCS6 A0A1Q3EXJ2 A0A1B6KS84 E2C3T7 A0A1B6DZD4 A0A0A1XA85 A0A1Y1M2V5 A0A182Y021 A0A336LQZ3
A0A0N0PAP9 A0A2H1VH86 A0A2J7QBL5 K7IM45 A0A232FG39 A0A1J1HS36 A0A088A1B9 A0A067QNE5 A0A0L7R5P8 A0A0M9A026 A0A0C9QWR0 A0A310SJF3 A0A2A3EM40 A0A1A9WEX0 A0A151I693 A0A154P6V9 A0A195FWM9 A0A195E3S2 A0A026WVV6 A0A151XA95 A0A158NNB7 F4WLG8 D6WKG0 A0A195AUI8 N6U0L2 A0A0J7P2B6 U4TY03 A0A2J7QBL1 A0A182JM71 A0A182HFI6 E9IUG5 A0A0K8W6D6 A0A182MRN2 A0A182P4V8 A0A1Y1M6F4 A0A084VND7 A0A0T6AVE5 A0A182F615 Q17JG4 A0A182W7A4 A0A182Q578 A0A182GSK8 E1ZZV0 A0A182RC82 A0A1W4WNU1 A0A182WU99 A0A182L1Q6 A0A182HUW7 B0W7M5 A0A2P8Z2J0 A0A182USE2 A0A1S4HDJ6 A0A182TIB1 A0A1B6FCS6 A0A1Q3EXJ2 A0A1B6KS84 E2C3T7 A0A1B6DZD4 A0A0A1XA85 A0A1Y1M2V5 A0A182Y021 A0A336LQZ3
Ontologies
PANTHER
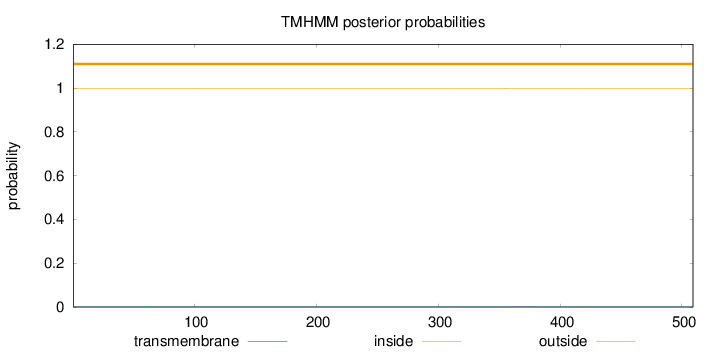
Topology
Length:
509
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01241
Exp number, first 60 AAs:
0.00022
Total prob of N-in:
0.00174
outside
1 - 509
Population Genetic Test Statistics
Pi
286.671115
Theta
173.89764
Tajima's D
2.100065
CLR
0.260394
CSRT
0.897755112244388
Interpretation
Uncertain
