Gene
KWMTBOMO09792
Pre Gene Modal
BGIBMGA009949
Annotation
PREDICTED:_uncharacterized_protein_LOC101741325_isoform_X1?_partial_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.176
Sequence
CDS
ATGACTGTGTGGTGGCTACAAATTCCCTTCGTCTACTGGATAACGGGAAGAAAAGCGACAGAGAATGCTTACGTTAAGAAAATAGACAGATTAACTTCGGATTTCTTGAAGAAGAGACGTACAGCTTTGAAGGGAGGTAATGTTGATGAAGAGTCAATGGGTATAGTCGATAGATACATTGCATCCGGTGAGCTGACAGAACAAGAAATCAAATGGGAGACGATGACTTTGTTTACAACGAGTCAAGAAGCTTCTGCAAAAATAACATCAGCTGTATTGTTATTTTTGGCGCACCTTCCTGACTGGCAGGAGAAAGTTTACAAGGAAATTGTCGAAGTAATCGGTTCAGGACGAAATGATGTTACAGCGGAACATTTAAAGCATCTACATTATTTAGATATGGTGTATCAAGAGGCACTGAGGTATCTGTCAATAGCAGCTCTGATACAAAGAACTGTGGAGGAAGAAATTACAATTAACAATGGAAAATTCACGCTTCCAATCGGTACGACATTAGTGATACCAATACACGACCTCCACAGAGACCCGAGGTATTGGGACGAGCCTTTGAAAGTTAAACCAGAGAGATTCCTCCCTGAAAATGTTAAGAAGAGGAGCCCTAATGTTTTTATTCCGTTCAGTTTAGGGGCGATGGACTGCTTAGGTCGAGTGTACGCAGAGCCGTTGATCAAAACGCTGGTAGTGTGGGCAGTGCGCGAAGTTCAGCTAGAAGCAGAAGGTTGCGTCGAAGACCTGAAACTACACGTCGCTATATCAGTGAAGTTCGCCAACGGATACAACTTAAAAGTCAAGCCAAGGATCTAA
Protein
MTVWWLQIPFVYWITGRKATENAYVKKIDRLTSDFLKKRRTALKGGNVDEESMGIVDRYIASGELTEQEIKWETMTLFTTSQEASAKITSAVLLFLAHLPDWQEKVYKEIVEVIGSGRNDVTAEHLKHLHYLDMVYQEALRYLSIAALIQRTVEEEITINNGKFTLPIGTTLVIPIHDLHRDPRYWDEPLKVKPERFLPENVKKRSPNVFIPFSLGAMDCLGRVYAEPLIKTLVVWAVREVQLEAEGCVEDLKLHVAISVKFANGYNLKVKPRI
Summary
Similarity
Belongs to the cytochrome P450 family.
Uniprot
H9JK98
H9LJ80
A0A2A4JUY6
A0A212FN17
J7FJY4
A0A2W1BU78
+ More
A0A068EWN3 A0A0K8TV51 A0A0K8TVP0 A0A2H1WX44 A0A248QHM1 A0A1V0D9J2 A0A411E587 X5D4I7 A0A437AW77 A0A2A4IZA0 A0A2H1WV90 A0A2W1C397 A0A248QG88 A0A068ETT0 A0A411E5J7 A0A0C5C1K7 J7FKN6 A0A3S2KZU1 A0A212FMZ1 A0A437B147 A0A1V0D9I7 A0A0K8TUH1 N6U8P7 I1VJ30 U4U9H7 A0A261BV48 X5DQS3 A0A1Y1N471 A0A261ADV0 A0A0C9PJ00 G5EC97 E3N782 A0A2G5THM7 A0A1Y1KEC4 A8Y4M2 A0A0K1YWG5 A0A090L5H4 V5K559 A0A0C9RFM1 A0A232FIT9 K7IZM3 A0A139WKY0 Q9U8F5 A0A1Y1KEU4 A0A454XVX9 H3G466 A0A1W6L1R4 A0A1Y1NMV2 G0N8C3 A0A0C9QZ23 A0A1J1HWV1 A0A0C9RC41 A0A162CMX4 A0A2G5TI30 A0A1Y1KCR3 K4JRV9 A0A1E1WM70 A0A0C9QJ49 A0A0K0LBC2 A0A2H2I8J5 E3MD49 A0A0K0LBC1 A0A1Y1NIE3 A0A1J1J298 A0A1Y1NLM3 A0A0K8TVT3 G0NPQ1 A0A1B6GEJ6 A0A0P5BZZ4 A0A0M3RSR5 A0A139WNP8 K7IVR3 A0A0P5W212 A0A0P5UL65 V5GQ09 A0A0P5V468 A0A0P5WNK8 A0A261BDQ0 A0A2P4WHR7 A0A067QR22 A0A0P4YSY3 A0A0N5BE60 A0A0P5V6E5 A0A2J7QZQ8 E3MZ92 A0A0N7ZP20 G0NPQ4 A0A226E8D8 V5HS62 A8XVD6 A0A2R5LK85
A0A068EWN3 A0A0K8TV51 A0A0K8TVP0 A0A2H1WX44 A0A248QHM1 A0A1V0D9J2 A0A411E587 X5D4I7 A0A437AW77 A0A2A4IZA0 A0A2H1WV90 A0A2W1C397 A0A248QG88 A0A068ETT0 A0A411E5J7 A0A0C5C1K7 J7FKN6 A0A3S2KZU1 A0A212FMZ1 A0A437B147 A0A1V0D9I7 A0A0K8TUH1 N6U8P7 I1VJ30 U4U9H7 A0A261BV48 X5DQS3 A0A1Y1N471 A0A261ADV0 A0A0C9PJ00 G5EC97 E3N782 A0A2G5THM7 A0A1Y1KEC4 A8Y4M2 A0A0K1YWG5 A0A090L5H4 V5K559 A0A0C9RFM1 A0A232FIT9 K7IZM3 A0A139WKY0 Q9U8F5 A0A1Y1KEU4 A0A454XVX9 H3G466 A0A1W6L1R4 A0A1Y1NMV2 G0N8C3 A0A0C9QZ23 A0A1J1HWV1 A0A0C9RC41 A0A162CMX4 A0A2G5TI30 A0A1Y1KCR3 K4JRV9 A0A1E1WM70 A0A0C9QJ49 A0A0K0LBC2 A0A2H2I8J5 E3MD49 A0A0K0LBC1 A0A1Y1NIE3 A0A1J1J298 A0A1Y1NLM3 A0A0K8TVT3 G0NPQ1 A0A1B6GEJ6 A0A0P5BZZ4 A0A0M3RSR5 A0A139WNP8 K7IVR3 A0A0P5W212 A0A0P5UL65 V5GQ09 A0A0P5V468 A0A0P5WNK8 A0A261BDQ0 A0A2P4WHR7 A0A067QR22 A0A0P4YSY3 A0A0N5BE60 A0A0P5V6E5 A0A2J7QZQ8 E3MZ92 A0A0N7ZP20 G0NPQ4 A0A226E8D8 V5HS62 A8XVD6 A0A2R5LK85
Pubmed
EMBL
BABH01042142
JF751066
AEF13244.1
NWSH01000564
PCG75619.1
AGBW02007649
+ More
OWR55127.1 JX310090 AFP20601.1 KZ149898 PZC78632.1 KM016719 AID54871.1 GCVX01000089 JAI18141.1 GCVX01000095 JAI18135.1 ODYU01011315 SOQ56984.1 KX443472 ASO98047.1 KY212092 ARA91656.1 MH236447 QBA57452.1 KF701176 AHW57346.1 RSAL01000342 RVE42386.1 NWSH01004296 PCG65257.1 SOQ56980.1 PZC78633.1 KX443473 ASO98048.1 KM016720 AID54872.1 MH236448 QBA57453.1 KP001146 AJN91191.1 JX310092 AFP20603.1 RVE42385.1 AGBW02007650 OWR55114.1 RSAL01000212 RVE44177.1 KY212093 ARA91657.1 GCVX01000091 JAI18139.1 APGK01035437 KB740925 ENN78040.1 JQ855651 AFI45014.1 KB631924 ERL87266.1 NIPN01000077 OZG13625.1 KF701178 AHW57348.1 GEZM01014048 JAV92208.1 NMWX01000007 OZF96130.1 GBYB01000938 JAG70705.1 BX284605 CAA98548.1 DS268546 EFO88453.1 PDUG01000005 PIC26768.1 GEZM01088768 JAV57955.1 HE601533 CAP39842.1 KP859404 AKZ17714.1 LN609528 CEF64982.1 KF044265 AGT57837.1 GBYB01007184 GBYB01007185 JAG76951.1 JAG76952.1 NNAY01000144 OXU30595.1 KQ971326 KYB28487.1 AF091117 AAF09264.1 GEZM01088778 GEZM01088777 GEZM01088771 JAV57936.1 ABKE03000024 PDM78918.1 KX609529 ARN17936.1 GEZM01001581 JAV97656.1 GL379850 EGT55334.1 GBYB01000975 JAG70742.1 CVRI01000020 CRK91006.1 GBYB01014154 JAG83921.1 LRGB01000944 KZS14706.1 PIC26918.1 GEZM01088761 JAV57970.1 JX876518 JX462987 AFU86448.1 AGN52780.1 GDQN01009628 GDQN01002954 JAT81426.1 JAT88100.1 GBYB01003574 JAG73341.1 KM217052 KM217053 KM217054 AIW80016.1 AIW80017.1 DS268436 EFO98909.1 KM217047 AIW80010.1 GEZM01001583 JAV97654.1 CVRI01000067 CRL06511.1 GEZM01001582 JAV97655.1 GCVX01000055 JAI18175.1 GL379921 EGT35305.1 GECZ01009036 JAS60733.1 GDIP01192928 JAJ30474.1 KR012836 ALD15911.1 KQ971311 KYB29496.1 GDIP01092738 JAM10977.1 GDIP01111344 JAL92370.1 GANP01012008 JAB72460.1 GDIP01120749 JAL82965.1 GDIP01204930 GDIP01179080 GDIP01084697 GDIP01042788 JAM19018.1 NMWX01000001 OZG08449.1 LFJK02000005 POM49085.1 KK853474 KDR07369.1 GDIP01224122 JAI99279.1 GDIP01104948 JAL98766.1 NEVH01009068 PNF34044.1 DS268500 EFP12903.1 GDIP01226577 JAI96824.1 EGT35361.1 LNIX01000005 OXA53364.1 GANP01005666 JAB78802.1 HE601047 CAP36603.1 GGLE01005763 MBY09889.1
OWR55127.1 JX310090 AFP20601.1 KZ149898 PZC78632.1 KM016719 AID54871.1 GCVX01000089 JAI18141.1 GCVX01000095 JAI18135.1 ODYU01011315 SOQ56984.1 KX443472 ASO98047.1 KY212092 ARA91656.1 MH236447 QBA57452.1 KF701176 AHW57346.1 RSAL01000342 RVE42386.1 NWSH01004296 PCG65257.1 SOQ56980.1 PZC78633.1 KX443473 ASO98048.1 KM016720 AID54872.1 MH236448 QBA57453.1 KP001146 AJN91191.1 JX310092 AFP20603.1 RVE42385.1 AGBW02007650 OWR55114.1 RSAL01000212 RVE44177.1 KY212093 ARA91657.1 GCVX01000091 JAI18139.1 APGK01035437 KB740925 ENN78040.1 JQ855651 AFI45014.1 KB631924 ERL87266.1 NIPN01000077 OZG13625.1 KF701178 AHW57348.1 GEZM01014048 JAV92208.1 NMWX01000007 OZF96130.1 GBYB01000938 JAG70705.1 BX284605 CAA98548.1 DS268546 EFO88453.1 PDUG01000005 PIC26768.1 GEZM01088768 JAV57955.1 HE601533 CAP39842.1 KP859404 AKZ17714.1 LN609528 CEF64982.1 KF044265 AGT57837.1 GBYB01007184 GBYB01007185 JAG76951.1 JAG76952.1 NNAY01000144 OXU30595.1 KQ971326 KYB28487.1 AF091117 AAF09264.1 GEZM01088778 GEZM01088777 GEZM01088771 JAV57936.1 ABKE03000024 PDM78918.1 KX609529 ARN17936.1 GEZM01001581 JAV97656.1 GL379850 EGT55334.1 GBYB01000975 JAG70742.1 CVRI01000020 CRK91006.1 GBYB01014154 JAG83921.1 LRGB01000944 KZS14706.1 PIC26918.1 GEZM01088761 JAV57970.1 JX876518 JX462987 AFU86448.1 AGN52780.1 GDQN01009628 GDQN01002954 JAT81426.1 JAT88100.1 GBYB01003574 JAG73341.1 KM217052 KM217053 KM217054 AIW80016.1 AIW80017.1 DS268436 EFO98909.1 KM217047 AIW80010.1 GEZM01001583 JAV97654.1 CVRI01000067 CRL06511.1 GEZM01001582 JAV97655.1 GCVX01000055 JAI18175.1 GL379921 EGT35305.1 GECZ01009036 JAS60733.1 GDIP01192928 JAJ30474.1 KR012836 ALD15911.1 KQ971311 KYB29496.1 GDIP01092738 JAM10977.1 GDIP01111344 JAL92370.1 GANP01012008 JAB72460.1 GDIP01120749 JAL82965.1 GDIP01204930 GDIP01179080 GDIP01084697 GDIP01042788 JAM19018.1 NMWX01000001 OZG08449.1 LFJK02000005 POM49085.1 KK853474 KDR07369.1 GDIP01224122 JAI99279.1 GDIP01104948 JAL98766.1 NEVH01009068 PNF34044.1 DS268500 EFP12903.1 GDIP01226577 JAI96824.1 EGT35361.1 LNIX01000005 OXA53364.1 GANP01005666 JAB78802.1 HE601047 CAP36603.1 GGLE01005763 MBY09889.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000019118
UP000030742
+ More
UP000216463 UP000216624 UP000001940 UP000008281 UP000230233 UP000008549 UP000035682 UP000215335 UP000002358 UP000007266 UP000232936 UP000005239 UP000008068 UP000183832 UP000076858 UP000005237 UP000237256 UP000027135 UP000046392 UP000235965 UP000198287
UP000216463 UP000216624 UP000001940 UP000008281 UP000230233 UP000008549 UP000035682 UP000215335 UP000002358 UP000007266 UP000232936 UP000005239 UP000008068 UP000183832 UP000076858 UP000005237 UP000237256 UP000027135 UP000046392 UP000235965 UP000198287
Interpro
Gene 3D
ProteinModelPortal
H9JK98
H9LJ80
A0A2A4JUY6
A0A212FN17
J7FJY4
A0A2W1BU78
+ More
A0A068EWN3 A0A0K8TV51 A0A0K8TVP0 A0A2H1WX44 A0A248QHM1 A0A1V0D9J2 A0A411E587 X5D4I7 A0A437AW77 A0A2A4IZA0 A0A2H1WV90 A0A2W1C397 A0A248QG88 A0A068ETT0 A0A411E5J7 A0A0C5C1K7 J7FKN6 A0A3S2KZU1 A0A212FMZ1 A0A437B147 A0A1V0D9I7 A0A0K8TUH1 N6U8P7 I1VJ30 U4U9H7 A0A261BV48 X5DQS3 A0A1Y1N471 A0A261ADV0 A0A0C9PJ00 G5EC97 E3N782 A0A2G5THM7 A0A1Y1KEC4 A8Y4M2 A0A0K1YWG5 A0A090L5H4 V5K559 A0A0C9RFM1 A0A232FIT9 K7IZM3 A0A139WKY0 Q9U8F5 A0A1Y1KEU4 A0A454XVX9 H3G466 A0A1W6L1R4 A0A1Y1NMV2 G0N8C3 A0A0C9QZ23 A0A1J1HWV1 A0A0C9RC41 A0A162CMX4 A0A2G5TI30 A0A1Y1KCR3 K4JRV9 A0A1E1WM70 A0A0C9QJ49 A0A0K0LBC2 A0A2H2I8J5 E3MD49 A0A0K0LBC1 A0A1Y1NIE3 A0A1J1J298 A0A1Y1NLM3 A0A0K8TVT3 G0NPQ1 A0A1B6GEJ6 A0A0P5BZZ4 A0A0M3RSR5 A0A139WNP8 K7IVR3 A0A0P5W212 A0A0P5UL65 V5GQ09 A0A0P5V468 A0A0P5WNK8 A0A261BDQ0 A0A2P4WHR7 A0A067QR22 A0A0P4YSY3 A0A0N5BE60 A0A0P5V6E5 A0A2J7QZQ8 E3MZ92 A0A0N7ZP20 G0NPQ4 A0A226E8D8 V5HS62 A8XVD6 A0A2R5LK85
A0A068EWN3 A0A0K8TV51 A0A0K8TVP0 A0A2H1WX44 A0A248QHM1 A0A1V0D9J2 A0A411E587 X5D4I7 A0A437AW77 A0A2A4IZA0 A0A2H1WV90 A0A2W1C397 A0A248QG88 A0A068ETT0 A0A411E5J7 A0A0C5C1K7 J7FKN6 A0A3S2KZU1 A0A212FMZ1 A0A437B147 A0A1V0D9I7 A0A0K8TUH1 N6U8P7 I1VJ30 U4U9H7 A0A261BV48 X5DQS3 A0A1Y1N471 A0A261ADV0 A0A0C9PJ00 G5EC97 E3N782 A0A2G5THM7 A0A1Y1KEC4 A8Y4M2 A0A0K1YWG5 A0A090L5H4 V5K559 A0A0C9RFM1 A0A232FIT9 K7IZM3 A0A139WKY0 Q9U8F5 A0A1Y1KEU4 A0A454XVX9 H3G466 A0A1W6L1R4 A0A1Y1NMV2 G0N8C3 A0A0C9QZ23 A0A1J1HWV1 A0A0C9RC41 A0A162CMX4 A0A2G5TI30 A0A1Y1KCR3 K4JRV9 A0A1E1WM70 A0A0C9QJ49 A0A0K0LBC2 A0A2H2I8J5 E3MD49 A0A0K0LBC1 A0A1Y1NIE3 A0A1J1J298 A0A1Y1NLM3 A0A0K8TVT3 G0NPQ1 A0A1B6GEJ6 A0A0P5BZZ4 A0A0M3RSR5 A0A139WNP8 K7IVR3 A0A0P5W212 A0A0P5UL65 V5GQ09 A0A0P5V468 A0A0P5WNK8 A0A261BDQ0 A0A2P4WHR7 A0A067QR22 A0A0P4YSY3 A0A0N5BE60 A0A0P5V6E5 A0A2J7QZQ8 E3MZ92 A0A0N7ZP20 G0NPQ4 A0A226E8D8 V5HS62 A8XVD6 A0A2R5LK85
PDB
6MA7
E-value=7.04052e-22,
Score=255
Ontologies
GO
Topology
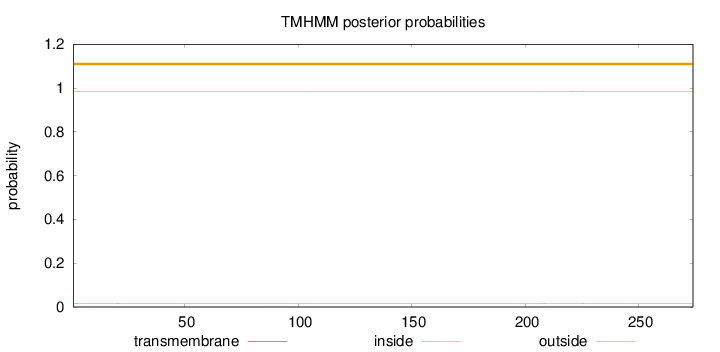
Length:
274
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01413
Exp number, first 60 AAs:
0.00345
Total prob of N-in:
0.01642
outside
1 - 274
Population Genetic Test Statistics
Pi
371.1827
Theta
189.581146
Tajima's D
1.895716
CLR
0.746222
CSRT
0.866706664666767
Interpretation
Uncertain
