Gene
KWMTBOMO09788 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013111
Annotation
cellular_retinoic_acid_binding_protein_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.476
Sequence
CDS
ATGGAATTCGTAGGCAAGAAATACAAGATGACTTCCTCCGAGAACTTTGATGAGTTCATGAAGACCATCGGCGTGGGTCTGATCACCCGCAAAGCCGCCAACGCGGTCACCCCGACCGTGGAGCTCCGTAAGGATGGTGACGAATACAATTTAGTCACCTCTTCAACCTTCAAGACCACTGAGATGAAGTTCAAGCCCGGCGAAGAGTTCGAAGAGGACCGCGCTGACGGTGCTAAGGTGAAATCCGTATGCACATTCGAAGGCAACACCCTCAAGCAAGTCCAGAAGGCCCCCGACGGTCTTGAAGTCACTTACGTCAGGGAATTCGGCCCTGAGGAGATGAAAGCTGTGATGACAGCTAAGGACGTGACTTGCACCAGAGTCTACAAGGTCCAGTAA
Protein
MEFVGKKYKMTSSENFDEFMKTIGVGLITRKAANAVTPTVELRKDGDEYNLVTSSTFKTTEMKFKPGEEFEEDRADGAKVKSVCTFEGNTLKQVQKAPDGLEVTYVREFGPEEMKAVMTAKDVTCTRVYKVQ
Summary
Similarity
Belongs to the calycin superfamily. Fatty-acid binding protein (FABP) family.
Uniprot
Q2QEH2
O61236
A0A212FN10
Q0WX97
M4M9L0
A0A0K8SYG9
+ More
B6CMG0 A0A2A4J1H5 A0A1E1WC74 A0A3G1T1H0 Q6F440 I4DJD9 I4DN38 A0A0A7BYM6 A0A0N1I4W9 A0A2A4AQU9 S4P8B4 A0A437BU53 A0A2H1V4G9 A0A0L7LQG2 A0A1W4WNF7 A0A195BSW7 A0A3L8D7Y7 F6MFX1 D5LXI1 V9IIS4 J3JTW6 A0A151XBY8 Q76LA4 A0A0L7R215 A0A139WGC8 F4WMC4 A0A0K8TL91 A0A2A3ERN5 A0A1W4WMG9 A0A088AE74 A0A158P294 A0A0J7L3Z2 K7IX95 A0A0N0U575 K7IX94 A0A0G3YKZ9 E2AWC1 E2BJH6 T1E383 A0A232EJI6 A0A411G7C2 D2A4Y9 R4V0J3 A0A1L8DZ33 A0A1B0EX54 R4WCU0 R4WCK5 A0A1B6KYE8 A0A0V0G2M9 T1HLW6 A0A1A9V645 A0A224XNU5 A0A0P4VYV4 B3P4J3 A0A023F8M4 A0A1B6DMS6 A0A0T6BBK9 A0A2S2PV04 A0A023EFS1 Q177Y4 T1DJZ8 A0A336N2B1 A0A182VXR6 A0A084WE60 A0A2M4AEI4 A0A2M4CU83 A0A182FP88 A0A2M3YZB1 A0A182PUC3 F5HLQ7 A0A1Q3F8S3 A0A1B6H6S1 A0A182IWT0 B0X309 A0A1B6GJK3 Q1M0X8 C4WXA0 T1J6K4 A0A2S2NIM3 A0A1B6LZC8 A0A195F346 Q8WR15 A0A443SL95 A0A1L8EC06 Q0PXX4 A0A1Y1NDG9 A0A026WE75 A0A336LH33 E9G4Z6 A0A1B6KEV6 A0A1S3DRQ9 A0A1I8M6S3
B6CMG0 A0A2A4J1H5 A0A1E1WC74 A0A3G1T1H0 Q6F440 I4DJD9 I4DN38 A0A0A7BYM6 A0A0N1I4W9 A0A2A4AQU9 S4P8B4 A0A437BU53 A0A2H1V4G9 A0A0L7LQG2 A0A1W4WNF7 A0A195BSW7 A0A3L8D7Y7 F6MFX1 D5LXI1 V9IIS4 J3JTW6 A0A151XBY8 Q76LA4 A0A0L7R215 A0A139WGC8 F4WMC4 A0A0K8TL91 A0A2A3ERN5 A0A1W4WMG9 A0A088AE74 A0A158P294 A0A0J7L3Z2 K7IX95 A0A0N0U575 K7IX94 A0A0G3YKZ9 E2AWC1 E2BJH6 T1E383 A0A232EJI6 A0A411G7C2 D2A4Y9 R4V0J3 A0A1L8DZ33 A0A1B0EX54 R4WCU0 R4WCK5 A0A1B6KYE8 A0A0V0G2M9 T1HLW6 A0A1A9V645 A0A224XNU5 A0A0P4VYV4 B3P4J3 A0A023F8M4 A0A1B6DMS6 A0A0T6BBK9 A0A2S2PV04 A0A023EFS1 Q177Y4 T1DJZ8 A0A336N2B1 A0A182VXR6 A0A084WE60 A0A2M4AEI4 A0A2M4CU83 A0A182FP88 A0A2M3YZB1 A0A182PUC3 F5HLQ7 A0A1Q3F8S3 A0A1B6H6S1 A0A182IWT0 B0X309 A0A1B6GJK3 Q1M0X8 C4WXA0 T1J6K4 A0A2S2NIM3 A0A1B6LZC8 A0A195F346 Q8WR15 A0A443SL95 A0A1L8EC06 Q0PXX4 A0A1Y1NDG9 A0A026WE75 A0A336LH33 E9G4Z6 A0A1B6KEV6 A0A1S3DRQ9 A0A1I8M6S3
Pubmed
17966127
9618497
22118469
18760362
28756777
22651552
+ More
26354079 23622113 26227816 30249741 22516182 23537049 14745104 18362917 19820115 21719571 26369729 21347285 20075255 25702953 20798317 24330624 28648823 23691247 27129103 17994087 25474469 24945155 26483478 17510324 24438588 12364791 14747013 17210077 12034496 28004739 24508170 21292972 25315136
26354079 23622113 26227816 30249741 22516182 23537049 14745104 18362917 19820115 21719571 26369729 21347285 20075255 25702953 20798317 24330624 28648823 23691247 27129103 17994087 25474469 24945155 26483478 17510324 24438588 12364791 14747013 17210077 12034496 28004739 24508170 21292972 25315136
EMBL
DQ109670
DQ311348
DQ539018
EF112401
AB262579
AAZ82096.1
+ More
ABD36292.1 ABG20590.1 ABK97427.1 BAE96009.1 U75307 AAC24317.1 AGBW02007650 OWR55117.1 AB267808 BAF02663.1 KC127672 AGG56524.1 GBRD01007489 JAG58332.1 EU325560 KZ149898 ACB54950.1 PZC78673.1 NWSH01003893 PCG65719.1 GDQN01006476 JAT84578.1 MG992392 AXY94830.1 AB180450 BAD26694.1 AK401407 KQ459582 BAM18029.1 KPI98807.1 AK402706 BAM19328.1 KC966936 AHV85210.1 KQ461183 KPJ07543.1 NWMT01000107 PCC99322.1 GAIX01006016 JAA86544.1 RSAL01000008 RVE53998.1 ODYU01000601 SOQ35676.1 JTDY01000327 KOB77682.1 KQ976418 KYM89450.1 QOIP01000012 RLU16018.1 JF815103 AEG74454.1 GU980922 ADE27971.1 JR049662 AEY61033.1 APGK01053973 BT126675 KB741240 KB632257 AEE61638.1 ENN72097.1 ERL90786.1 KQ982315 KYQ57859.1 AB083010 BAC54131.1 KQ414666 KOC64879.1 KQ971345 KYB27002.1 GL888217 EGI64768.1 GDAI01002486 JAI15117.1 KZ288192 PBC34398.1 ADTU01001164 LBMM01000937 KMQ97189.1 AAZX01000617 AAZX01000618 KQ435794 KOX73671.1 KM507112 AKM28436.1 GL443280 EFN62281.1 GL448571 EFN84133.1 GALA01000600 JAA94252.1 NNAY01003987 OXU18530.1 MH366001 QBB01810.1 EFA05702.1 KC632308 AGM32122.1 GFDF01002361 JAV11723.1 AJWK01030958 AJWK01030959 AJWK01030960 AJWK01030961 AK416989 BAN20204.1 AK416948 BAN20163.1 GEBQ01023515 JAT16462.1 GECL01003738 JAP02386.1 ACPB03014789 GFTR01002259 JAW14167.1 GDKW01001862 JAI54733.1 CH954181 EDV49508.1 GBBI01000966 JAC17746.1 GEDC01010368 JAS26930.1 LJIG01002210 KRT84730.1 GGMS01000088 MBY69291.1 JXUM01012309 JXUM01012310 GAPW01005897 KQ560351 JAC07701.1 KXJ82893.1 CH477369 EAT42473.1 GAMD01001196 JAB00395.1 UFQT01006868 SSX36141.1 ATLV01023157 KE525341 KFB48504.1 GGFK01005717 MBW39038.1 GGFL01004726 MBW68904.1 GGFM01000835 MBW21586.1 AAAB01008904 EGK97217.1 EGK97218.1 GFDL01011093 JAV23952.1 GECU01037290 JAS70416.1 DS233316 DS232304 EDS29395.1 EDS39511.1 GECZ01014401 GECZ01007162 JAS55368.1 JAS62607.1 AY792955 AAX33735.1 ABLF02030056 AK342527 BAH72520.1 JH431878 GGMR01004411 MBY17030.1 GEBQ01010987 JAT28990.1 KQ981855 KYN34806.1 AF458289 AAL68638.1 NCKV01001465 RWS28297.1 GFDG01002529 JAV16270.1 DQ673412 ABG81985.1 GEZM01006856 JAV95618.1 KK107261 EZA53971.1 UFQS01003764 UFQT01003764 SSX15925.1 SSX35267.1 GL732532 EFX85386.1 GEBQ01029975 JAT10002.1
ABD36292.1 ABG20590.1 ABK97427.1 BAE96009.1 U75307 AAC24317.1 AGBW02007650 OWR55117.1 AB267808 BAF02663.1 KC127672 AGG56524.1 GBRD01007489 JAG58332.1 EU325560 KZ149898 ACB54950.1 PZC78673.1 NWSH01003893 PCG65719.1 GDQN01006476 JAT84578.1 MG992392 AXY94830.1 AB180450 BAD26694.1 AK401407 KQ459582 BAM18029.1 KPI98807.1 AK402706 BAM19328.1 KC966936 AHV85210.1 KQ461183 KPJ07543.1 NWMT01000107 PCC99322.1 GAIX01006016 JAA86544.1 RSAL01000008 RVE53998.1 ODYU01000601 SOQ35676.1 JTDY01000327 KOB77682.1 KQ976418 KYM89450.1 QOIP01000012 RLU16018.1 JF815103 AEG74454.1 GU980922 ADE27971.1 JR049662 AEY61033.1 APGK01053973 BT126675 KB741240 KB632257 AEE61638.1 ENN72097.1 ERL90786.1 KQ982315 KYQ57859.1 AB083010 BAC54131.1 KQ414666 KOC64879.1 KQ971345 KYB27002.1 GL888217 EGI64768.1 GDAI01002486 JAI15117.1 KZ288192 PBC34398.1 ADTU01001164 LBMM01000937 KMQ97189.1 AAZX01000617 AAZX01000618 KQ435794 KOX73671.1 KM507112 AKM28436.1 GL443280 EFN62281.1 GL448571 EFN84133.1 GALA01000600 JAA94252.1 NNAY01003987 OXU18530.1 MH366001 QBB01810.1 EFA05702.1 KC632308 AGM32122.1 GFDF01002361 JAV11723.1 AJWK01030958 AJWK01030959 AJWK01030960 AJWK01030961 AK416989 BAN20204.1 AK416948 BAN20163.1 GEBQ01023515 JAT16462.1 GECL01003738 JAP02386.1 ACPB03014789 GFTR01002259 JAW14167.1 GDKW01001862 JAI54733.1 CH954181 EDV49508.1 GBBI01000966 JAC17746.1 GEDC01010368 JAS26930.1 LJIG01002210 KRT84730.1 GGMS01000088 MBY69291.1 JXUM01012309 JXUM01012310 GAPW01005897 KQ560351 JAC07701.1 KXJ82893.1 CH477369 EAT42473.1 GAMD01001196 JAB00395.1 UFQT01006868 SSX36141.1 ATLV01023157 KE525341 KFB48504.1 GGFK01005717 MBW39038.1 GGFL01004726 MBW68904.1 GGFM01000835 MBW21586.1 AAAB01008904 EGK97217.1 EGK97218.1 GFDL01011093 JAV23952.1 GECU01037290 JAS70416.1 DS233316 DS232304 EDS29395.1 EDS39511.1 GECZ01014401 GECZ01007162 JAS55368.1 JAS62607.1 AY792955 AAX33735.1 ABLF02030056 AK342527 BAH72520.1 JH431878 GGMR01004411 MBY17030.1 GEBQ01010987 JAT28990.1 KQ981855 KYN34806.1 AF458289 AAL68638.1 NCKV01001465 RWS28297.1 GFDG01002529 JAV16270.1 DQ673412 ABG81985.1 GEZM01006856 JAV95618.1 KK107261 EZA53971.1 UFQS01003764 UFQT01003764 SSX15925.1 SSX35267.1 GL732532 EFX85386.1 GEBQ01029975 JAT10002.1
Proteomes
UP000007151
UP000218220
UP000053268
UP000053240
UP000243750
UP000283053
+ More
UP000037510 UP000192223 UP000078540 UP000279307 UP000019118 UP000030742 UP000075809 UP000053825 UP000007266 UP000007755 UP000242457 UP000005203 UP000005205 UP000036403 UP000002358 UP000053105 UP000000311 UP000008237 UP000215335 UP000092461 UP000015103 UP000078200 UP000008711 UP000069940 UP000249989 UP000008820 UP000075920 UP000030765 UP000069272 UP000075885 UP000007062 UP000075880 UP000002320 UP000007819 UP000078541 UP000288716 UP000079169 UP000053097 UP000000305 UP000095301
UP000037510 UP000192223 UP000078540 UP000279307 UP000019118 UP000030742 UP000075809 UP000053825 UP000007266 UP000007755 UP000242457 UP000005203 UP000005205 UP000036403 UP000002358 UP000053105 UP000000311 UP000008237 UP000215335 UP000092461 UP000015103 UP000078200 UP000008711 UP000069940 UP000249989 UP000008820 UP000075920 UP000030765 UP000069272 UP000075885 UP000007062 UP000075880 UP000002320 UP000007819 UP000078541 UP000288716 UP000079169 UP000053097 UP000000305 UP000095301
Pfam
PF00061 Lipocalin
Interpro
SUPFAM
SSF50814
SSF50814
Gene 3D
ProteinModelPortal
Q2QEH2
O61236
A0A212FN10
Q0WX97
M4M9L0
A0A0K8SYG9
+ More
B6CMG0 A0A2A4J1H5 A0A1E1WC74 A0A3G1T1H0 Q6F440 I4DJD9 I4DN38 A0A0A7BYM6 A0A0N1I4W9 A0A2A4AQU9 S4P8B4 A0A437BU53 A0A2H1V4G9 A0A0L7LQG2 A0A1W4WNF7 A0A195BSW7 A0A3L8D7Y7 F6MFX1 D5LXI1 V9IIS4 J3JTW6 A0A151XBY8 Q76LA4 A0A0L7R215 A0A139WGC8 F4WMC4 A0A0K8TL91 A0A2A3ERN5 A0A1W4WMG9 A0A088AE74 A0A158P294 A0A0J7L3Z2 K7IX95 A0A0N0U575 K7IX94 A0A0G3YKZ9 E2AWC1 E2BJH6 T1E383 A0A232EJI6 A0A411G7C2 D2A4Y9 R4V0J3 A0A1L8DZ33 A0A1B0EX54 R4WCU0 R4WCK5 A0A1B6KYE8 A0A0V0G2M9 T1HLW6 A0A1A9V645 A0A224XNU5 A0A0P4VYV4 B3P4J3 A0A023F8M4 A0A1B6DMS6 A0A0T6BBK9 A0A2S2PV04 A0A023EFS1 Q177Y4 T1DJZ8 A0A336N2B1 A0A182VXR6 A0A084WE60 A0A2M4AEI4 A0A2M4CU83 A0A182FP88 A0A2M3YZB1 A0A182PUC3 F5HLQ7 A0A1Q3F8S3 A0A1B6H6S1 A0A182IWT0 B0X309 A0A1B6GJK3 Q1M0X8 C4WXA0 T1J6K4 A0A2S2NIM3 A0A1B6LZC8 A0A195F346 Q8WR15 A0A443SL95 A0A1L8EC06 Q0PXX4 A0A1Y1NDG9 A0A026WE75 A0A336LH33 E9G4Z6 A0A1B6KEV6 A0A1S3DRQ9 A0A1I8M6S3
B6CMG0 A0A2A4J1H5 A0A1E1WC74 A0A3G1T1H0 Q6F440 I4DJD9 I4DN38 A0A0A7BYM6 A0A0N1I4W9 A0A2A4AQU9 S4P8B4 A0A437BU53 A0A2H1V4G9 A0A0L7LQG2 A0A1W4WNF7 A0A195BSW7 A0A3L8D7Y7 F6MFX1 D5LXI1 V9IIS4 J3JTW6 A0A151XBY8 Q76LA4 A0A0L7R215 A0A139WGC8 F4WMC4 A0A0K8TL91 A0A2A3ERN5 A0A1W4WMG9 A0A088AE74 A0A158P294 A0A0J7L3Z2 K7IX95 A0A0N0U575 K7IX94 A0A0G3YKZ9 E2AWC1 E2BJH6 T1E383 A0A232EJI6 A0A411G7C2 D2A4Y9 R4V0J3 A0A1L8DZ33 A0A1B0EX54 R4WCU0 R4WCK5 A0A1B6KYE8 A0A0V0G2M9 T1HLW6 A0A1A9V645 A0A224XNU5 A0A0P4VYV4 B3P4J3 A0A023F8M4 A0A1B6DMS6 A0A0T6BBK9 A0A2S2PV04 A0A023EFS1 Q177Y4 T1DJZ8 A0A336N2B1 A0A182VXR6 A0A084WE60 A0A2M4AEI4 A0A2M4CU83 A0A182FP88 A0A2M3YZB1 A0A182PUC3 F5HLQ7 A0A1Q3F8S3 A0A1B6H6S1 A0A182IWT0 B0X309 A0A1B6GJK3 Q1M0X8 C4WXA0 T1J6K4 A0A2S2NIM3 A0A1B6LZC8 A0A195F346 Q8WR15 A0A443SL95 A0A1L8EC06 Q0PXX4 A0A1Y1NDG9 A0A026WE75 A0A336LH33 E9G4Z6 A0A1B6KEV6 A0A1S3DRQ9 A0A1I8M6S3
PDB
3PPT
E-value=1.80225e-25,
Score=281
Ontologies
GO
PANTHER
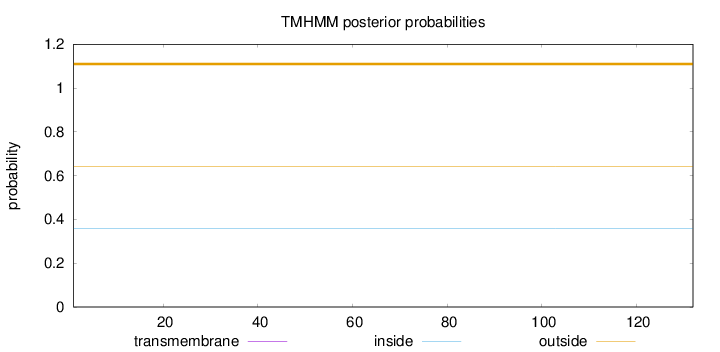
Topology
Length:
132
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00055
Exp number, first 60 AAs:
0.00055
Total prob of N-in:
0.35821
outside
1 - 132
Population Genetic Test Statistics
Pi
250.208437
Theta
184.114904
Tajima's D
1.455159
CLR
0.343012
CSRT
0.781010949452527
Interpretation
Uncertain
