Gene
KWMTBOMO09779
Annotation
GMP_synthase_[Operophtera_brumata]
Location in the cell
Nuclear Reliability : 4.003
Sequence
CDS
ATGCCCGCAGTGAAAGTATTCAACATATTCGGCGGGCAGAAGCCGACCGAGAAGAACCCGCCGGCCTCACCCCCCTCGCAACAGGGAGCCGCCCAAAAGTTCTACAGGACTTTTTCAGTCGCTCCAGAAGTCTCGGTCGAGCACAAGGGCTCGAAAGTGACCGCGATAGCCGCTTGCCTAGAAAAGAAAATAAGTGAGTCCCCCGTGAAAGTCAGCAGTGAGGTGAAAATTACTAAAAACAATGTTTGTTGTTCCGGTGTAAAAGATAAGCCTAAAGCGAGAGCGCGTCAACGCGTATACGAGAATGTAAACATAGTGACGGTGCCTGGTAATGAAACAAATGACGACAGACTACTTGAAGAAAATAAAGGATACTGTTCGATTTATGAAAACGTGACAGTTGTGAACAAACAGCTTGATCATCGTGTGTCCGAAACGAAACACATTTCTGAATCTAAACTTACTAGAGCTTTGGAATCGTTCGATAAATTACTGTCTGAGTTCTCATCAAACATTTCGTTGAACTCTGAACTGAAATCAGTTAAAAGTGAATTTAATCCGCCTAAACTTCAAAAATCTAAAACGTGTAGTATCATCGAATCTCATTGCATTTTAAAGAAAGCCAATTCTGATCCTGAAAACACTAAAACTCGAAGTCGTGTCTCCAGGAATAATTCCATAGAAAAAACTACTAGTCTTTGGAACTTAGACGATATGAAGGGAAAAGAGTTAAGTGCTCCACTGGTACCTCTGTCCCCTAAAACTAACGACAAGATGAAACCAGATAAATATGCTACGTATAAAGTTTCTTCAAAGGGCGTATCACGGTCAAACTGTCTTAAAACTGAAGATTTAAAAAAACTTGACGTTAAGACAAGAATACTCAAGAAAACCCTATCTAATCCACCGTCGACACCCGTCCCGGTTGTAACTAAAACGAAGCCGCTGACAAAAAAAGTCGAGAAAAAAGTGAACGACCCAAAGAAAATAAAACCCAAAACGTGTACAGATAATGGATCGTTAAATATGAAGACGAATTTAGAAAGTAAAACGCCCCAAAAATCACAAATGCAGAAAGCCAAAAGTGTGTGGGAAATAGGAAATGAAATGGCTCTAATTACCCCTATTCTAGAAAGGACAAAATCTACGACTTCTATCACGGGTTCTCCTAGTAAGATCCCGGTGATTAGGAGTCAAATATCGCTCAACAAGTTCAGTAGTACCAGAGCTCTATTTTCTCCCACTCCGGTTGATCTATATCAAGTAGACCAGGCCAAGGAGTGTGCTCAGAAAAAGAAACCGATTACCGCACGGAAGGCAACGGAAAAGTCAGATAACTTTAATACAAAACAGGTCAGACAAAAATCACAAGTCAACATCAAAGTTGAAGGAAAACGACAAAGCGAGAAAATTAAAAAAGATACCGACGTCAAAAAACTGGTTTCGTCTCCGAAATCATTAAGGAAAGATTATACAGACGAAATTAATGCAATACGAGTAAAACTGCAGCAGAAAAAAAACAATAAAAGAGATCTTATCGTTGTTGTGCCGGATGCCAAACGCAACGACAATAAACCGCTCGGAATTGATTCGACATTGTCGCCCGTCAAGTCGATCGTAAAAAAGCTGGAGCTGAAGACTGCTTCGGAGTTGAACACAGATCAGAATCAGTTCACGAATTGTAAAGTCATACCGCCGGTGCACAAAGAAGTATGCGTGGCGAATACAACATTCCACAATCACTTGAGCACACTCGTAGGCCGCCAAATCAAATACGTAGAGTCGAGAGATGATTCTAAAACGTGCTCTCAAGTCGAGAAGCTTACATTAAACAAAGACTGCGATGAGAAGATATCAGATACGCACTCAGACTGCAGCGATGACTCGGGTCACGTTTCCAACGACGCTGCGAATGATAACGAACAGACTTTCGATAACGTACGCGATGTGAGCCCGGTAGTGATTAGCGTCGACGAGGTGGATTGCAAACCTTTCGACGGACCCCAACAGTTTGGGATAGATTTGTCTGAGAATGCCAAGACTGTGTGTCCCGTGCGGCCGGCGAGGCGCTGCGGGCGCTCCAACGAGGTCGCTAGCGGTACCCACGCCACCGACACTCCGAAAGCGGACGAACAGGTACGCGACACTATTAACATATCGATTATTATTATCGATAACAACAAATTACAGGTGCCGGCTACAGCCCGCCGGTGTCGACATGCGGTCATTCGGATCGCAACAGATACTAAATCTTACGGCCACTATATCAACTAG
Protein
MPAVKVFNIFGGQKPTEKNPPASPPSQQGAAQKFYRTFSVAPEVSVEHKGSKVTAIAACLEKKISESPVKVSSEVKITKNNVCCSGVKDKPKARARQRVYENVNIVTVPGNETNDDRLLEENKGYCSIYENVTVVNKQLDHRVSETKHISESKLTRALESFDKLLSEFSSNISLNSELKSVKSEFNPPKLQKSKTCSIIESHCILKKANSDPENTKTRSRVSRNNSIEKTTSLWNLDDMKGKELSAPLVPLSPKTNDKMKPDKYATYKVSSKGVSRSNCLKTEDLKKLDVKTRILKKTLSNPPSTPVPVVTKTKPLTKKVEKKVNDPKKIKPKTCTDNGSLNMKTNLESKTPQKSQMQKAKSVWEIGNEMALITPILERTKSTTSITGSPSKIPVIRSQISLNKFSSTRALFSPTPVDLYQVDQAKECAQKKKPITARKATEKSDNFNTKQVRQKSQVNIKVEGKRQSEKIKKDTDVKKLVSSPKSLRKDYTDEINAIRVKLQQKKNNKRDLIVVVPDAKRNDNKPLGIDSTLSPVKSIVKKLELKTASELNTDQNQFTNCKVIPPVHKEVCVANTTFHNHLSTLVGRQIKYVESRDDSKTCSQVEKLTLNKDCDEKISDTHSDCSDDSGHVSNDAANDNEQTFDNVRDVSPVVISVDEVDCKPFDGPQQFGIDLSENAKTVCPVRPARRCGRSNEVASGTHATDTPKADEQVRDTINISIIIIDNNKLQVPATARRCRHAVIRIATDTKSYGHYIN
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
Uniprot
EMBL
Proteomes
Pfam
PF00063 Myosin_head
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
Ontologies
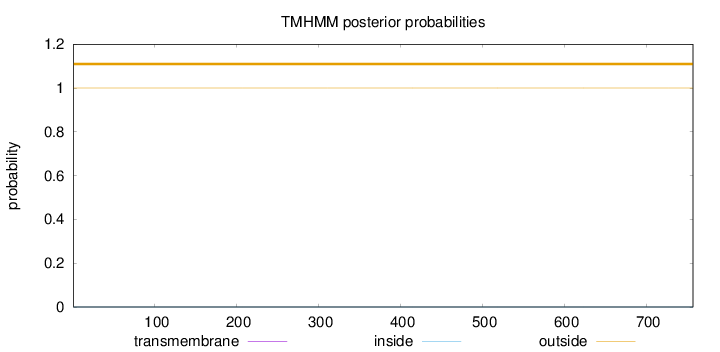
Topology
Length:
757
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00004
outside
1 - 757
Population Genetic Test Statistics
Pi
306.585994
Theta
201.022367
Tajima's D
1.348826
CLR
0
CSRT
0.752012399380031
Interpretation
Uncertain
