Pre Gene Modal
BGIBMGA012921
Annotation
PREDICTED:_unconventional_myosin-XVIIIa?_partial_[Bombyx_mori]
Location in the cell
Cytoskeletal Reliability : 1.127 Mitochondrial Reliability : 1.293
Sequence
CDS
ATGTTCAGGGGCGGCGTCGTCGGCGGCTTGGACGCGCGTCGAGACGCCGCGCTTGCCCGCGTGCTCGTTCGCCTGCAGGCGAGGGCGCGCGGGTTGCTCGCCCGACGCAAGGCCCAGCGTTTGCGGACGCAGCACACGGCCGCACGGTGCATCCAACGCAACGTGCGGGCCTTCCTCACGGTGCGGGACTGGCCTTGGTGGAGGCTGCTCGTGCGAGTCACGCCCCTGCTCGCCGTGCACAGGACTGAACATCGACTTAAACAAGCACAGGACGAGTTGGAGACGCTTCGTTCTAAATTGGACAAAGTCGAAAACGAAAGGACACATTACAAGAGCGAAACCGAACGACTCGAGACTAAGCTCTCGGAAGTGGGAGGTGAGCTAGCAGAGGAAAGAGCGGCCGCCGGACTCGCAACGGAACGAGCGGACGCCGAGGCTGCAGAACGCCTTCGAGTGGAGAAGGAGAACAGAGACTTGGTCGCCAATAACCAGCGATTACAGCAGGCTTCAGAACGTCTAGAACTTGAACTCTTGCATTGGCGTTCGGCCGATAACGGAGCAGCGGAACATTCAGATTCGGAGGGATCCGAAGCGGACGCGGCCTCCGCCGGCGACAAGTACAAACGGCGATACGAACGCGCCCACCGAGAGCTGCAGCTGATCAGGGCGCAATTGAGACGCCAACACGAGGACGACCTCGAACAACTCGTTACGGTCAAGAAGCAGTTGGAGAAGAAGGTTCAAGATGCATACGAAGAGGTCGAAGAACAGCGAGCCGTGGCCGCGCAATGGAAACGAAAATTGCAGAAACTCACCAACGACATGGCCGACCTGAGGTCGCTTCTCGACGAACAGACGTGCCGCAACAACTTGCTGGAGAAACGCCAGCGTAAGTTCGACGGTGAGCTACAGGCGGCGCAGGAAGAACTGAAGAGAGAACGATCGGCCAAGGAGCGGATCTCTCGCGAGAGAGATCTAGCGCAGGCGGAGAAGTTCGCAATGGAGCAGTCCCTTTCTGAAGCTCGTATGGAGTTGGAGCTGAAAGAAGAGCGGCTGTCGGCGGCGGCGCGCGAGCTGGAGGAGCGGGGCGGCGGCGGCGACGAGCTGGCGGCGCTGCGCCGCGCCCGCGCCGACCTCGAGCGCCGCGTGCGCGACCAGGACGAGGAGCTCGACGACCTCGCCGGACAGATACAGTTGCTCGAAAGTTCGAAACTTCGCCTGGAAATGTTACTGGAGCAGCAGCGCAAGGAAGCACGCCTGGAAGCCGCCGCCCGCGACGACGAAATGGAGGAGACCAGAGCCAACGCTGTCAAGAAACTGAAGATACTGGAGAGTCAGCTGGAGAGCGAGCACGCCGAGCGCAGCCTGCTGCTCCGCGAGCGCCACGAGCTGGAGCGGCGCCTGGCCGCGCTCGAGGAGGCCGCGCGCGCCGACACGCACGAGCACGCGCAGCTCGCGCAGAGACTCAAGCGGTACGTACCTAACTAA
Protein
MFRGGVVGGLDARRDAALARVLVRLQARARGLLARRKAQRLRTQHTAARCIQRNVRAFLTVRDWPWWRLLVRVTPLLAVHRTEHRLKQAQDELETLRSKLDKVENERTHYKSETERLETKLSEVGGELAEERAAAGLATERADAEAAERLRVEKENRDLVANNQRLQQASERLELELLHWRSADNGAAEHSDSEGSEADAASAGDKYKRRYERAHRELQLIRAQLRRQHEDDLEQLVTVKKQLEKKVQDAYEEVEEQRAVAAQWKRKLQKLTNDMADLRSLLDEQTCRNNLLEKRQRKFDGELQAAQEELKRERSAKERISRERDLAQAEKFAMEQSLSEARMELELKEERLSAAARELEERGGGGDELAALRRARADLERRVRDQDEELDDLAGQIQLLESSKLRLEMLLEQQRKEARLEAAARDDEMEETRANAVKKLKILESQLESEHAERSLLLRERHELERRLAALEEAARADTHEHAQLAQRLKRYVPN
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
Uniprot
A0A2W1BYD3
A0A3S2M8N9
A0A2J7RBE9
A0A2J7RBF5
A0A067RC42
A0A139WGL1
+ More
A0A0R1E367 K7J5Q2 F4WIA4 A0A1W4VP06 Q8IH49 A0A151WPF3 A0A336KMA4 A0A1Y1KQA3 A0A1W4WCA9 A0A1Y1KMY5 A0A1L8E548 A0A1L8E4V2 A0A1L8E4V9 A0A195F260 A0A158P0H4 A0A1W4VBY6 A0A0Q9XA56 A0A1W4VQA3 B4KAF0 A0A0Q9XAP8 A0A348G644 A0A0Q9X279 A0A0Q9XAY5 A0A0Q9X0J5 A0A0P8YPG9 A0A1W4VBW5 B3LZY8 A0A0Q9X0N3 A0A0P9A230 A0A0N8P1R5 A0A0R3NJN7 A0A0R3NPQ9 Q294V0 B4NL11 A0A0R3NJI5 I5APN9 B3P0Y0 A0A0R3NJQ4 A0A3B0JN63 A0A0R1E726 A0A0R1E3R7 B4PR41 A0A0R1E337 A0A0R1E331 Q9VEZ0 Q8INC3 Q7K523 A0A0A1X1N6 Q0KI67 Q0KI66 A0A0Q9WQD2 A0A0Q9WNE5 B4LY10 A0A0Q9WFE7 T1PBT3 A0A0Q9WEF7 A0A0Q9WEQ3 T1PMH2 A0A0Q9WPK8 T1PKM2 A0A1I8PN66 B4QXQ6 A0A1I8PN26 A0A1I8PN68 B4JG99 A0A1I8PN10 A0A1I8N1D3 T1PBP0 A0A1Q3EU29 A0A0K8UTA8 A0A1Q3FUB1 A0A0K8VQN6 Q5TVZ3 E0W3P1 A0A2H8TUT0 J9JNI5 A0A2H4N1N4 A0A2S2P795 A0A1B6KRK5 A0A423TMF8 A0A1B6LX51 A0A1B6EBK3 A0A1B6CI70 A0A2S2Q1Q8
A0A0R1E367 K7J5Q2 F4WIA4 A0A1W4VP06 Q8IH49 A0A151WPF3 A0A336KMA4 A0A1Y1KQA3 A0A1W4WCA9 A0A1Y1KMY5 A0A1L8E548 A0A1L8E4V2 A0A1L8E4V9 A0A195F260 A0A158P0H4 A0A1W4VBY6 A0A0Q9XA56 A0A1W4VQA3 B4KAF0 A0A0Q9XAP8 A0A348G644 A0A0Q9X279 A0A0Q9XAY5 A0A0Q9X0J5 A0A0P8YPG9 A0A1W4VBW5 B3LZY8 A0A0Q9X0N3 A0A0P9A230 A0A0N8P1R5 A0A0R3NJN7 A0A0R3NPQ9 Q294V0 B4NL11 A0A0R3NJI5 I5APN9 B3P0Y0 A0A0R3NJQ4 A0A3B0JN63 A0A0R1E726 A0A0R1E3R7 B4PR41 A0A0R1E337 A0A0R1E331 Q9VEZ0 Q8INC3 Q7K523 A0A0A1X1N6 Q0KI67 Q0KI66 A0A0Q9WQD2 A0A0Q9WNE5 B4LY10 A0A0Q9WFE7 T1PBT3 A0A0Q9WEF7 A0A0Q9WEQ3 T1PMH2 A0A0Q9WPK8 T1PKM2 A0A1I8PN66 B4QXQ6 A0A1I8PN26 A0A1I8PN68 B4JG99 A0A1I8PN10 A0A1I8N1D3 T1PBP0 A0A1Q3EU29 A0A0K8UTA8 A0A1Q3FUB1 A0A0K8VQN6 Q5TVZ3 E0W3P1 A0A2H8TUT0 J9JNI5 A0A2H4N1N4 A0A2S2P795 A0A1B6KRK5 A0A423TMF8 A0A1B6LX51 A0A1B6EBK3 A0A1B6CI70 A0A2S2Q1Q8
Pubmed
EMBL
KZ149898
PZC78665.1
RSAL01000008
RVE54001.1
NEVH01005904
PNF38164.1
+ More
PNF38166.1 KK852802 KDR16277.1 KQ971345 KYB27001.1 CM000160 KRK03677.1 GL888175 EGI66035.1 AE014297 BT001428 AAN13696.2 AAN71183.1 KQ982860 KYQ49779.1 UFQS01000594 UFQT01000594 SSX05202.1 SSX25563.1 GEZM01078315 JAV62768.1 GEZM01078314 JAV62769.1 GFDF01000320 JAV13764.1 GFDF01000321 JAV13763.1 GFDF01000314 JAV13770.1 KQ981856 KYN34665.1 ADTU01004978 ADTU01004979 ADTU01004980 CH933806 KRG01666.1 EDW15663.2 KRG01662.1 FX985584 BBF97917.1 CH964272 KRF99993.1 KRG01665.1 KRG01663.1 CH902617 KPU80655.1 EDV44178.2 KRG01664.1 KPU80657.1 KPU80661.1 KPU80660.1 CM000070 KRT01127.1 KRT01133.1 EAL28864.3 EDW84214.2 KRT01128.1 EIM52924.2 KRT01129.1 CH954181 EDV49099.1 KRT01131.1 OUUW01000008 SPP83665.1 KRK03681.1 KRK03679.1 EDW97363.2 KRK03675.1 KRK03680.1 AAF55271.3 AAN13695.2 AY051503 AAK92927.1 GBXI01009048 JAD05244.1 ABI31171.1 ABI31172.1 CH940650 KRF82958.1 KRF82961.1 EDW66876.2 KRF82962.1 KA645570 AFP60199.1 KRF82957.1 KRF82960.1 KRF82963.1 KA649295 AFP63924.1 KRF82959.1 KA649296 AFP63925.1 CM000364 EDX12730.1 CH916369 EDV92568.1 KA645525 AFP60154.1 GFDL01016238 JAV18807.1 GDHF01033592 GDHF01022385 GDHF01020550 JAI18722.1 JAI29929.1 JAI31764.1 GFDL01003979 JAV31066.1 GDHF01011439 GDHF01001328 JAI40875.1 JAI50986.1 AAAB01008816 EAL41598.3 DS235882 EEB20247.1 GFXV01005895 MBW17700.1 ABLF02037475 ABLF02037488 KY653069 ATU47263.1 GGMR01012710 MBY25329.1 GEBQ01025927 JAT14050.1 QCYY01001502 ROT77628.1 GEBQ01011746 JAT28231.1 GEDC01002005 JAS35293.1 GEDC01024273 JAS13025.1 GGMS01002495 MBY71698.1
PNF38166.1 KK852802 KDR16277.1 KQ971345 KYB27001.1 CM000160 KRK03677.1 GL888175 EGI66035.1 AE014297 BT001428 AAN13696.2 AAN71183.1 KQ982860 KYQ49779.1 UFQS01000594 UFQT01000594 SSX05202.1 SSX25563.1 GEZM01078315 JAV62768.1 GEZM01078314 JAV62769.1 GFDF01000320 JAV13764.1 GFDF01000321 JAV13763.1 GFDF01000314 JAV13770.1 KQ981856 KYN34665.1 ADTU01004978 ADTU01004979 ADTU01004980 CH933806 KRG01666.1 EDW15663.2 KRG01662.1 FX985584 BBF97917.1 CH964272 KRF99993.1 KRG01665.1 KRG01663.1 CH902617 KPU80655.1 EDV44178.2 KRG01664.1 KPU80657.1 KPU80661.1 KPU80660.1 CM000070 KRT01127.1 KRT01133.1 EAL28864.3 EDW84214.2 KRT01128.1 EIM52924.2 KRT01129.1 CH954181 EDV49099.1 KRT01131.1 OUUW01000008 SPP83665.1 KRK03681.1 KRK03679.1 EDW97363.2 KRK03675.1 KRK03680.1 AAF55271.3 AAN13695.2 AY051503 AAK92927.1 GBXI01009048 JAD05244.1 ABI31171.1 ABI31172.1 CH940650 KRF82958.1 KRF82961.1 EDW66876.2 KRF82962.1 KA645570 AFP60199.1 KRF82957.1 KRF82960.1 KRF82963.1 KA649295 AFP63924.1 KRF82959.1 KA649296 AFP63925.1 CM000364 EDX12730.1 CH916369 EDV92568.1 KA645525 AFP60154.1 GFDL01016238 JAV18807.1 GDHF01033592 GDHF01022385 GDHF01020550 JAI18722.1 JAI29929.1 JAI31764.1 GFDL01003979 JAV31066.1 GDHF01011439 GDHF01001328 JAI40875.1 JAI50986.1 AAAB01008816 EAL41598.3 DS235882 EEB20247.1 GFXV01005895 MBW17700.1 ABLF02037475 ABLF02037488 KY653069 ATU47263.1 GGMR01012710 MBY25329.1 GEBQ01025927 JAT14050.1 QCYY01001502 ROT77628.1 GEBQ01011746 JAT28231.1 GEDC01002005 JAS35293.1 GEDC01024273 JAS13025.1 GGMS01002495 MBY71698.1
Proteomes
UP000283053
UP000235965
UP000027135
UP000007266
UP000002282
UP000002358
+ More
UP000007755 UP000192221 UP000000803 UP000075809 UP000192223 UP000078541 UP000005205 UP000009192 UP000007798 UP000007801 UP000001819 UP000008711 UP000268350 UP000008792 UP000095300 UP000000304 UP000001070 UP000095301 UP000007062 UP000009046 UP000007819 UP000283509
UP000007755 UP000192221 UP000000803 UP000075809 UP000192223 UP000078541 UP000005205 UP000009192 UP000007798 UP000007801 UP000001819 UP000008711 UP000268350 UP000008792 UP000095300 UP000000304 UP000001070 UP000095301 UP000007062 UP000009046 UP000007819 UP000283509
Interpro
Gene 3D
ProteinModelPortal
A0A2W1BYD3
A0A3S2M8N9
A0A2J7RBE9
A0A2J7RBF5
A0A067RC42
A0A139WGL1
+ More
A0A0R1E367 K7J5Q2 F4WIA4 A0A1W4VP06 Q8IH49 A0A151WPF3 A0A336KMA4 A0A1Y1KQA3 A0A1W4WCA9 A0A1Y1KMY5 A0A1L8E548 A0A1L8E4V2 A0A1L8E4V9 A0A195F260 A0A158P0H4 A0A1W4VBY6 A0A0Q9XA56 A0A1W4VQA3 B4KAF0 A0A0Q9XAP8 A0A348G644 A0A0Q9X279 A0A0Q9XAY5 A0A0Q9X0J5 A0A0P8YPG9 A0A1W4VBW5 B3LZY8 A0A0Q9X0N3 A0A0P9A230 A0A0N8P1R5 A0A0R3NJN7 A0A0R3NPQ9 Q294V0 B4NL11 A0A0R3NJI5 I5APN9 B3P0Y0 A0A0R3NJQ4 A0A3B0JN63 A0A0R1E726 A0A0R1E3R7 B4PR41 A0A0R1E337 A0A0R1E331 Q9VEZ0 Q8INC3 Q7K523 A0A0A1X1N6 Q0KI67 Q0KI66 A0A0Q9WQD2 A0A0Q9WNE5 B4LY10 A0A0Q9WFE7 T1PBT3 A0A0Q9WEF7 A0A0Q9WEQ3 T1PMH2 A0A0Q9WPK8 T1PKM2 A0A1I8PN66 B4QXQ6 A0A1I8PN26 A0A1I8PN68 B4JG99 A0A1I8PN10 A0A1I8N1D3 T1PBP0 A0A1Q3EU29 A0A0K8UTA8 A0A1Q3FUB1 A0A0K8VQN6 Q5TVZ3 E0W3P1 A0A2H8TUT0 J9JNI5 A0A2H4N1N4 A0A2S2P795 A0A1B6KRK5 A0A423TMF8 A0A1B6LX51 A0A1B6EBK3 A0A1B6CI70 A0A2S2Q1Q8
A0A0R1E367 K7J5Q2 F4WIA4 A0A1W4VP06 Q8IH49 A0A151WPF3 A0A336KMA4 A0A1Y1KQA3 A0A1W4WCA9 A0A1Y1KMY5 A0A1L8E548 A0A1L8E4V2 A0A1L8E4V9 A0A195F260 A0A158P0H4 A0A1W4VBY6 A0A0Q9XA56 A0A1W4VQA3 B4KAF0 A0A0Q9XAP8 A0A348G644 A0A0Q9X279 A0A0Q9XAY5 A0A0Q9X0J5 A0A0P8YPG9 A0A1W4VBW5 B3LZY8 A0A0Q9X0N3 A0A0P9A230 A0A0N8P1R5 A0A0R3NJN7 A0A0R3NPQ9 Q294V0 B4NL11 A0A0R3NJI5 I5APN9 B3P0Y0 A0A0R3NJQ4 A0A3B0JN63 A0A0R1E726 A0A0R1E3R7 B4PR41 A0A0R1E337 A0A0R1E331 Q9VEZ0 Q8INC3 Q7K523 A0A0A1X1N6 Q0KI67 Q0KI66 A0A0Q9WQD2 A0A0Q9WNE5 B4LY10 A0A0Q9WFE7 T1PBT3 A0A0Q9WEF7 A0A0Q9WEQ3 T1PMH2 A0A0Q9WPK8 T1PKM2 A0A1I8PN66 B4QXQ6 A0A1I8PN26 A0A1I8PN68 B4JG99 A0A1I8PN10 A0A1I8N1D3 T1PBP0 A0A1Q3EU29 A0A0K8UTA8 A0A1Q3FUB1 A0A0K8VQN6 Q5TVZ3 E0W3P1 A0A2H8TUT0 J9JNI5 A0A2H4N1N4 A0A2S2P795 A0A1B6KRK5 A0A423TMF8 A0A1B6LX51 A0A1B6EBK3 A0A1B6CI70 A0A2S2Q1Q8
PDB
3J04
E-value=8.8002e-08,
Score=136
Ontologies
GO
Topology
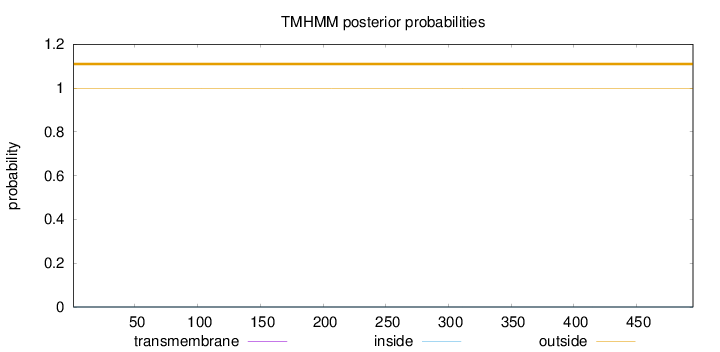
Length:
495
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00017
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00046
outside
1 - 495
Population Genetic Test Statistics
Pi
301.333016
Theta
172.487539
Tajima's D
2.44844
CLR
0.291447
CSRT
0.938603069846508
Interpretation
Uncertain
