Gene
KWMTBOMO09776 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012922
Annotation
PREDICTED:_AP-1_complex_subunit_beta-1_[Papilio_xuthus]
Full name
AP complex subunit beta
+ More
Beta-adaptin-like protein
Beta-adaptin-like protein
Location in the cell
Golgi Reliability : 2.517
Sequence
CDS
ATGACTGATTCCAAGTACTTCACCACTACGAAGAAAGGAGAGATATTCGAGTTGAAATCGGAACTAAACTCTGACAAAAAAGAGAAAAAGAAGGAGGCCGTTAAAAAGGTTATAGCATCCATGACAGTTGGAAAAGACGTCTCAGCATTGTTCCCCGATGTCGTGAATTGCATGCAGACAGATAACTTAGAACTGAAAAAACTGGTGTACTTATATTTAATGAATTATGCAAAGTCACAGCCAGACATGGCTATCATGGCGGTCAATACTTTTGTCAAGGACTGTGAGGATTCCAATCCATTGATTCGAGCTTTGGCTGTACGGACTATGGGATGCATTCGAGTGGACAAGATTACAGAATATCTATGCGAACCATTAAGAAAGTGCCTGAAAGATGAAGATCCTTATGTCCGGAAGACTGCAGCCGTGTGCGTCGCAAAGTTATATGATATCTCTTCAAGCATGGTTGAAGATCAGGGCTTCTTAGATCAACTAAAAGATCTGTTGAGCGATTCGAATCCTATGGTAGTAGCAAACGCTGTGGCAGCCTTATCAGAAATCAACGAAGCTAGTGTTTCTGGACGGCCCCTTGTTGAAATGAATGCCCCAACGATCAACAAGCTGTTGACTGCGTTGAATGAGTGTACTGAATGGGGTCAGGTGTTCATCCTGGACGCGTTGTCCAACTACTCGCCGCGTGACGCCCGCGAGGCGCACTCCATCTGCGAGAGGATCACGCCCCGCCTGGCGCACGCCAACGCCGCCGTCGTGCTCTCCGCTGTCAAGGTGCTGATGAAGCTGATGGAGATGGTGTCGGCGGACACGGAGCTGGTGAGCACGCTGTCCCGCAAGCTGGCGCCGCCGCTGGTGACGCTGCTGAGCGCGGAGCCCGAGGTGCAGTACGTGGCGCTGCGGAACATCAACCTCGTCGTGCAGAAGCGACCAGACATACTCAAACACGAGATGAAGGTGTTCTTTGTGAAGTACAATGACCCGATCTACGTTAAACTGGAGAAACTGGACATAATGATCCGATTGGCTTCCCAAGCCAACATAGCCCAGGTGCTCGGGGAACTGAAGGAGTATGCCACCGAAGTGGACGTGGACTTCGTCCGGAAGGCCGTCAGGGCTATAGGCAGATGCGCCATCAAGGTAGAGCCGTCGGCGGAGCGTTGCGTCTCTACATTACTAGAGCTGATCCAGACCAAAGTGAACTACGTCGTGCAAGAGGCCATTGTGGTGATCAAAGACATATTCAGAAAGTACCCGAACAAGTATGAGAGCATCATCAGTACGCTGTGCGAAAATCTTGATACTTTAGACGAGCCTGAGGCTAGGGCTTCCATGGTATGGATTGTGGGCGAATACGCGGAACGTATCGACAACGCAGACGAACTTCTAGATTCGTTCCTCGAAGGGTTCCACGATGAAAACGCACAGGTACAACTGCAACTCCTCACGGCCGTTGTGAAACTGTTCCTGAAACGTCCAGCCGACACGCAAGAGCTAGTACAGCACGTGCTTAGTCTGGCAACACAGGATTCGGACAATCCCGACTTGAGAGACCGCGGCTTCATCTACTGGCGTCTACTGTCGACCGACCCGGCCGCGGCCAAGGAAGTGGTCCTGGCCGACAAGCCTCTCATATCGGAGGAGACTGATCTTCTGGAACCCACACTACTGGACGAACTCATCTGCCACATCAGCTCGCTGGCTTCGGTCTATCACAAGCCACCGACGGCATTCGTTGAAGGCCGCGGCGCCGGAGTCCGCAAGTCCCTGCCGTCCCGAGGTGTTGTCGCCGAAGAAACGAACACACAACAACCCACAGTCATCCCGAATCAGGAGTCGTTGATCGGCGATCTATTGTCTATGGACATCGGAGGTCCCCCCCCGGCCCCGGCGCCCTCTTCCAACGTCGACCTCCTCGCCGGCGGACTAGATGTTCTCTTGGGCGGCGGGCCGGAGCCGGGCGCGTCCGCCGCGGCGGTCCCCGGCGGCGCCTCCGGCCTGCTCGGCGACATCTTCGCCGTCTCCGCCGCGCCCGCCTCCTACGTGCCGCCCAAACAGTGCTGGCTGCCCGCCGACAAAGGAAAAGGTTTGGAGATTTGGGGTACGTTCAGCCGTCAGAACGGTCAGCCTCGGATGGAGATGACATTCACTAACAAGGCGATGCAGCCGATGACAGGTTTCGCCATACAACTCAATAAGAACAGTTTCGGCGTTTATCCTGTGGGCGGGCTGGCGGTGGGCGCGGTGGGGGCGGGCGCGGGCGCGGAGGCCGCACTGCCGCTCGCCACGCACGGGCCCGTGCAACGCATGGACCCGCTCAACAACCTACAGGTCGCGATAAAGAACAATATAGATGTATTCTACTTCGCCTGCCTAATCCCCGCTCACATATTATTCACGGAAGACGGACAGCTAGACAAACGTGTGTTCCTCACGACTTGGAAGGAAATACCCGCAGCGAACGAAGTACAACATACACTATCGAACGTTGTGGGAACAGCCGATTCTATCGCTCAAAAAATGACGCTAAACAATATCTTTACTATCGCTAAACGTAATGTCGAAGGTCAAGATATGTTATATCAATCATTGAAGTTAACTAACAACATCTGGGTGTTGCTAGAGTTGAAACTTCAGCCCGGCAACCCCGAAGCGACGCTCAGCTTGAAGTCCCGAACTGTAGAGGTCGCCACGTGTATATTCCAAGCCTACGAAGCTATAATCAAATCGTAA
Protein
MTDSKYFTTTKKGEIFELKSELNSDKKEKKKEAVKKVIASMTVGKDVSALFPDVVNCMQTDNLELKKLVYLYLMNYAKSQPDMAIMAVNTFVKDCEDSNPLIRALAVRTMGCIRVDKITEYLCEPLRKCLKDEDPYVRKTAAVCVAKLYDISSSMVEDQGFLDQLKDLLSDSNPMVVANAVAALSEINEASVSGRPLVEMNAPTINKLLTALNECTEWGQVFILDALSNYSPRDAREAHSICERITPRLAHANAAVVLSAVKVLMKLMEMVSADTELVSTLSRKLAPPLVTLLSAEPEVQYVALRNINLVVQKRPDILKHEMKVFFVKYNDPIYVKLEKLDIMIRLASQANIAQVLGELKEYATEVDVDFVRKAVRAIGRCAIKVEPSAERCVSTLLELIQTKVNYVVQEAIVVIKDIFRKYPNKYESIISTLCENLDTLDEPEARASMVWIVGEYAERIDNADELLDSFLEGFHDENAQVQLQLLTAVVKLFLKRPADTQELVQHVLSLATQDSDNPDLRDRGFIYWRLLSTDPAAAKEVVLADKPLISEETDLLEPTLLDELICHISSLASVYHKPPTAFVEGRGAGVRKSLPSRGVVAEETNTQQPTVIPNQESLIGDLLSMDIGGPPPAPAPSSNVDLLAGGLDVLLGGGPEPGASAAAVPGGASGLLGDIFAVSAAPASYVPPKQCWLPADKGKGLEIWGTFSRQNGQPRMEMTFTNKAMQPMTGFAIQLNKNSFGVYPVGGLAVGAVGAGAGAEAALPLATHGPVQRMDPLNNLQVAIKNNIDVFYFACLIPAHILFTEDGQLDKRVFLTTWKEIPAANEVQHTLSNVVGTADSIAQKMTLNNIFTIAKRNVEGQDMLYQSLKLTNNIWVLLELKLQPGNPEATLSLKSRTVEVATCIFQAYEAIIKS
Summary
Description
Subunit of clathrin-associated adaptor protein complex that plays a role in protein sorting in the late-Golgi/trans-Golgi network (TGN) and/or endosomes. The AP complexes mediate both the recruitment of clathrin to membranes and the recognition of sorting signals within the cytosolic tails of transmembrane cargo molecules.
Subunit
Adaptor protein complexes are heterotetramers composed of two large adaptins (beta-type subunit and alpha-type or delta-type or epsilon-type or gamma-type subunit), a medium adaptin (mu-type subunit) and a small adaptin (sigma-type subunit).
Similarity
Belongs to the adaptor complexes large subunit family.
Uniprot
A0A437BU43
A0A2W1BYJ0
A0A2A4J8Z2
A0A0N1I9A1
A0A212FMC3
B4MA28
+ More
A0A0K8TY54 A0A034VKS9 B3NVT4 A0A1B0CED0 B4PZ07 A0A0A1WPY7 W8C5S6 B4JWU0 A0A1A9WDQ1 A0A1L8DVN8 B4NQ71 A0A0L0CKZ8 A0A0M4EWR3 B4L6V2 A0A1B0FI36 A0A1A9USV6 A0A1B0BB68 A0A1A9ZQI5 A0A1A9XCV0 A0A1Q3FY77 A0A1Y1LRA6 A0A1Q3FYB2 A0A1I8M3S5 A0A1I8NU89 A0A1W4XVD0 V5I8R8 U5EX18 E9J8K4 A0A158NBU8 K7ISE6 A0A2J7PX15 A0A1B6GJW3 A0A1B6IZT0 E2ACH8 A0A195BF10 A0A0C9QZS2 A0A067R983 A0A0A9Y0B4 A0A0C9R328 N6UAJ9 A0A069DWU5 A0A023F5S2 A0A224XE67 A0A0P4W4K3 A0A2P8ZJQ9 E0VK25 A0A0J7KLI9 A0A1B6DGS2 A0A0L7QWP9 A0A088AH86 A0A2G3AQQ1 F4X887 A0A195FDW1 V9I9B6 A0A2S2P3D1 A0A0N0BFV5 J9K6T5 A0A2A3ETU6 A0A195C9K4 A0A1B6BYD6 V9IAN3 A0A0P5P0E4 A0A0P6FYT9 A0A0P6EL28 A0A0P5MTA5 A0A0P6DXK2 A0A0P6G5U3 A0A0P5QZM6 A0A0P5RT68 A0A0P5K670 A0A0P5JVD7 A0A0P5RYG8 A0A0P5PUF0 A0A0N8C123 A0A0P5NT51 A0A0P6ET47 A0A0N8BXF5 A0A0P6DRX0 A0A0P5J4G0 A0A0P5KBT6 A0A151X9P3 A0A0P6G9T0 A0A0P5HDM1 A0A0P5S4Q2 A0A0P6ELN1 A0A0P5N8H0 A0A151J959 A0A0N8DUR1 A0A0N8C786 A0A0P6HE64 A0A3S3S074 A0A0K2UUH0 A0A132A7P0 A0A0P5BVX6
A0A0K8TY54 A0A034VKS9 B3NVT4 A0A1B0CED0 B4PZ07 A0A0A1WPY7 W8C5S6 B4JWU0 A0A1A9WDQ1 A0A1L8DVN8 B4NQ71 A0A0L0CKZ8 A0A0M4EWR3 B4L6V2 A0A1B0FI36 A0A1A9USV6 A0A1B0BB68 A0A1A9ZQI5 A0A1A9XCV0 A0A1Q3FY77 A0A1Y1LRA6 A0A1Q3FYB2 A0A1I8M3S5 A0A1I8NU89 A0A1W4XVD0 V5I8R8 U5EX18 E9J8K4 A0A158NBU8 K7ISE6 A0A2J7PX15 A0A1B6GJW3 A0A1B6IZT0 E2ACH8 A0A195BF10 A0A0C9QZS2 A0A067R983 A0A0A9Y0B4 A0A0C9R328 N6UAJ9 A0A069DWU5 A0A023F5S2 A0A224XE67 A0A0P4W4K3 A0A2P8ZJQ9 E0VK25 A0A0J7KLI9 A0A1B6DGS2 A0A0L7QWP9 A0A088AH86 A0A2G3AQQ1 F4X887 A0A195FDW1 V9I9B6 A0A2S2P3D1 A0A0N0BFV5 J9K6T5 A0A2A3ETU6 A0A195C9K4 A0A1B6BYD6 V9IAN3 A0A0P5P0E4 A0A0P6FYT9 A0A0P6EL28 A0A0P5MTA5 A0A0P6DXK2 A0A0P6G5U3 A0A0P5QZM6 A0A0P5RT68 A0A0P5K670 A0A0P5JVD7 A0A0P5RYG8 A0A0P5PUF0 A0A0N8C123 A0A0P5NT51 A0A0P6ET47 A0A0N8BXF5 A0A0P6DRX0 A0A0P5J4G0 A0A0P5KBT6 A0A151X9P3 A0A0P6G9T0 A0A0P5HDM1 A0A0P5S4Q2 A0A0P6ELN1 A0A0P5N8H0 A0A151J959 A0A0N8DUR1 A0A0N8C786 A0A0P6HE64 A0A3S3S074 A0A0K2UUH0 A0A132A7P0 A0A0P5BVX6
Pubmed
EMBL
RSAL01000008
RVE54002.1
KZ149898
PZC78664.1
NWSH01002600
PCG67882.1
+ More
KQ460635 KPJ13437.1 AGBW02007651 OWR54895.1 CH940655 EDW66087.1 KRF82552.1 GDHF01033299 GDHF01025458 JAI19015.1 JAI26856.1 GAKP01015031 JAC43921.1 CH954180 EDV46749.1 AJWK01008737 CM000162 EDX02085.1 GBXI01013400 JAD00892.1 GAMC01001552 JAC05004.1 CH916376 EDV95216.1 GFDF01003587 JAV10497.1 CH964291 EDW86296.1 JRES01000256 KNC32921.1 CP012528 ALC49947.1 CH933812 EDW06098.1 CCAG010004713 JXJN01011309 GFDL01002520 JAV32525.1 GEZM01054557 JAV73557.1 GFDL01002514 JAV32531.1 GALX01004210 JAB64256.1 GANO01001165 JAB58706.1 GL769034 EFZ10863.1 ADTU01011328 ADTU01011329 NEVH01020862 PNF20877.1 GECZ01007088 JAS62681.1 GECU01022471 GECU01015297 JAS85235.1 JAS92409.1 GL438525 EFN68864.1 KQ976500 KYM83163.1 GBYB01001215 JAG70982.1 KK852619 KDR20112.1 GBHO01018015 GBRD01016397 GDHC01002092 JAG25589.1 JAG49429.1 JAQ16537.1 GBYB01001216 JAG70983.1 APGK01036711 KB740941 ENN77661.1 GBGD01000356 JAC88533.1 GBBI01002190 JAC16522.1 GFTR01008338 JAW08088.1 GDKW01000326 JAI56269.1 PYGN01000035 PSN56732.1 DS235237 EEB13731.1 LBMM01005718 KMQ91273.1 GEDC01012407 JAS24891.1 KQ414713 KOC62964.1 MCIT02003576 PHT96513.1 GL888932 EGI57163.1 KQ981673 KYN38392.1 JR037303 JR037304 AEY57698.1 AEY57699.1 GGMR01011265 MBY23884.1 KQ435794 KOX73888.1 ABLF02024989 KZ288189 PBC34636.1 KQ978068 KYM97544.1 GEDC01031006 JAS06292.1 JR037301 JR037302 AEY57696.1 AEY57697.1 GDIQ01135418 JAL16308.1 GDIQ01058846 JAN35891.1 GDIQ01061047 JAN33690.1 GDIQ01151517 JAL00209.1 GDIQ01244794 GDIQ01169655 GDIQ01117434 GDIQ01079279 GDIQ01071690 LRGB01001005 JAN23047.1 KZS14136.1 GDIQ01038007 JAN56730.1 GDIQ01107972 JAL43754.1 GDIQ01096479 JAL55247.1 GDIQ01194599 JAK57126.1 GDIQ01194598 JAK57127.1 GDIQ01112374 GDIQ01112373 JAL39353.1 GDIQ01123220 JAL28506.1 GDIQ01125275 JAL26451.1 GDIQ01138125 JAL13601.1 GDIQ01071691 JAN23046.1 GDIQ01135419 JAL16307.1 GDIQ01074537 JAN20200.1 GDIQ01211087 JAK40638.1 GDIQ01192798 JAK58927.1 KQ982373 KYQ57039.1 GDIQ01038008 JAN56729.1 GDIQ01231383 JAK20342.1 GDIQ01092438 JAL59288.1 GDIQ01074536 JAN20201.1 GDIQ01156900 JAK94825.1 KQ979433 KYN21576.1 GDIQ01089980 JAN04757.1 GDIQ01107971 JAL43755.1 GDIQ01021271 JAN73466.1 NCKU01002919 RWS08542.1 HACA01024503 CDW41864.1 JXLN01011010 KPM06655.1 GDIP01179824 JAJ43578.1
KQ460635 KPJ13437.1 AGBW02007651 OWR54895.1 CH940655 EDW66087.1 KRF82552.1 GDHF01033299 GDHF01025458 JAI19015.1 JAI26856.1 GAKP01015031 JAC43921.1 CH954180 EDV46749.1 AJWK01008737 CM000162 EDX02085.1 GBXI01013400 JAD00892.1 GAMC01001552 JAC05004.1 CH916376 EDV95216.1 GFDF01003587 JAV10497.1 CH964291 EDW86296.1 JRES01000256 KNC32921.1 CP012528 ALC49947.1 CH933812 EDW06098.1 CCAG010004713 JXJN01011309 GFDL01002520 JAV32525.1 GEZM01054557 JAV73557.1 GFDL01002514 JAV32531.1 GALX01004210 JAB64256.1 GANO01001165 JAB58706.1 GL769034 EFZ10863.1 ADTU01011328 ADTU01011329 NEVH01020862 PNF20877.1 GECZ01007088 JAS62681.1 GECU01022471 GECU01015297 JAS85235.1 JAS92409.1 GL438525 EFN68864.1 KQ976500 KYM83163.1 GBYB01001215 JAG70982.1 KK852619 KDR20112.1 GBHO01018015 GBRD01016397 GDHC01002092 JAG25589.1 JAG49429.1 JAQ16537.1 GBYB01001216 JAG70983.1 APGK01036711 KB740941 ENN77661.1 GBGD01000356 JAC88533.1 GBBI01002190 JAC16522.1 GFTR01008338 JAW08088.1 GDKW01000326 JAI56269.1 PYGN01000035 PSN56732.1 DS235237 EEB13731.1 LBMM01005718 KMQ91273.1 GEDC01012407 JAS24891.1 KQ414713 KOC62964.1 MCIT02003576 PHT96513.1 GL888932 EGI57163.1 KQ981673 KYN38392.1 JR037303 JR037304 AEY57698.1 AEY57699.1 GGMR01011265 MBY23884.1 KQ435794 KOX73888.1 ABLF02024989 KZ288189 PBC34636.1 KQ978068 KYM97544.1 GEDC01031006 JAS06292.1 JR037301 JR037302 AEY57696.1 AEY57697.1 GDIQ01135418 JAL16308.1 GDIQ01058846 JAN35891.1 GDIQ01061047 JAN33690.1 GDIQ01151517 JAL00209.1 GDIQ01244794 GDIQ01169655 GDIQ01117434 GDIQ01079279 GDIQ01071690 LRGB01001005 JAN23047.1 KZS14136.1 GDIQ01038007 JAN56730.1 GDIQ01107972 JAL43754.1 GDIQ01096479 JAL55247.1 GDIQ01194599 JAK57126.1 GDIQ01194598 JAK57127.1 GDIQ01112374 GDIQ01112373 JAL39353.1 GDIQ01123220 JAL28506.1 GDIQ01125275 JAL26451.1 GDIQ01138125 JAL13601.1 GDIQ01071691 JAN23046.1 GDIQ01135419 JAL16307.1 GDIQ01074537 JAN20200.1 GDIQ01211087 JAK40638.1 GDIQ01192798 JAK58927.1 KQ982373 KYQ57039.1 GDIQ01038008 JAN56729.1 GDIQ01231383 JAK20342.1 GDIQ01092438 JAL59288.1 GDIQ01074536 JAN20201.1 GDIQ01156900 JAK94825.1 KQ979433 KYN21576.1 GDIQ01089980 JAN04757.1 GDIQ01107971 JAL43755.1 GDIQ01021271 JAN73466.1 NCKU01002919 RWS08542.1 HACA01024503 CDW41864.1 JXLN01011010 KPM06655.1 GDIP01179824 JAJ43578.1
Proteomes
UP000283053
UP000218220
UP000053240
UP000007151
UP000008792
UP000008711
+ More
UP000092461 UP000002282 UP000001070 UP000091820 UP000007798 UP000037069 UP000092553 UP000009192 UP000092444 UP000078200 UP000092460 UP000092445 UP000092443 UP000095301 UP000095300 UP000192223 UP000005205 UP000002358 UP000235965 UP000000311 UP000078540 UP000027135 UP000019118 UP000245037 UP000009046 UP000036403 UP000053825 UP000005203 UP000224522 UP000007755 UP000078541 UP000053105 UP000007819 UP000242457 UP000078542 UP000076858 UP000075809 UP000078492 UP000285301
UP000092461 UP000002282 UP000001070 UP000091820 UP000007798 UP000037069 UP000092553 UP000009192 UP000092444 UP000078200 UP000092460 UP000092445 UP000092443 UP000095301 UP000095300 UP000192223 UP000005205 UP000002358 UP000235965 UP000000311 UP000078540 UP000027135 UP000019118 UP000245037 UP000009046 UP000036403 UP000053825 UP000005203 UP000224522 UP000007755 UP000078541 UP000053105 UP000007819 UP000242457 UP000078542 UP000076858 UP000075809 UP000078492 UP000285301
Pfam
Interpro
IPR026739
AP_beta
+ More
IPR012295 TBP_dom_sf
IPR013037 Clathrin_b-adaptin_app_Ig-like
IPR016024 ARM-type_fold
IPR011989 ARM-like
IPR008152 Clathrin_a/b/g-adaptin_app_Ig
IPR009028 Coatomer/calthrin_app_sub_C
IPR016342 AP_complex_bsu_1_2_4
IPR015151 B-adaptin_app_sub_C
IPR002553 Clathrin/coatomer_adapt-like_N
IPR013041 Clathrin_app_Ig-like_sf
IPR022096 SBF1/SBF2
IPR037516 Tripartite_DENN
IPR001849 PH_domain
IPR001194 cDENN_dom
IPR004182 GRAM
IPR005113 uDENN_dom
IPR010569 Myotubularin-like_Pase_dom
IPR030564 Myotubularin_fam
IPR029021 Prot-tyrosine_phosphatase-like
IPR011993 PH-like_dom_sf
IPR005112 dDENN_dom
IPR002219 PE/DAG-bd
IPR036196 Ptyr_pPase_sf
IPR023485 Ptyr_pPase
IPR017867 Tyr_phospatase_low_mol_wt
IPR000225 Armadillo
IPR012295 TBP_dom_sf
IPR013037 Clathrin_b-adaptin_app_Ig-like
IPR016024 ARM-type_fold
IPR011989 ARM-like
IPR008152 Clathrin_a/b/g-adaptin_app_Ig
IPR009028 Coatomer/calthrin_app_sub_C
IPR016342 AP_complex_bsu_1_2_4
IPR015151 B-adaptin_app_sub_C
IPR002553 Clathrin/coatomer_adapt-like_N
IPR013041 Clathrin_app_Ig-like_sf
IPR022096 SBF1/SBF2
IPR037516 Tripartite_DENN
IPR001849 PH_domain
IPR001194 cDENN_dom
IPR004182 GRAM
IPR005113 uDENN_dom
IPR010569 Myotubularin-like_Pase_dom
IPR030564 Myotubularin_fam
IPR029021 Prot-tyrosine_phosphatase-like
IPR011993 PH-like_dom_sf
IPR005112 dDENN_dom
IPR002219 PE/DAG-bd
IPR036196 Ptyr_pPase_sf
IPR023485 Ptyr_pPase
IPR017867 Tyr_phospatase_low_mol_wt
IPR000225 Armadillo
SUPFAM
Gene 3D
CDD
ProteinModelPortal
A0A437BU43
A0A2W1BYJ0
A0A2A4J8Z2
A0A0N1I9A1
A0A212FMC3
B4MA28
+ More
A0A0K8TY54 A0A034VKS9 B3NVT4 A0A1B0CED0 B4PZ07 A0A0A1WPY7 W8C5S6 B4JWU0 A0A1A9WDQ1 A0A1L8DVN8 B4NQ71 A0A0L0CKZ8 A0A0M4EWR3 B4L6V2 A0A1B0FI36 A0A1A9USV6 A0A1B0BB68 A0A1A9ZQI5 A0A1A9XCV0 A0A1Q3FY77 A0A1Y1LRA6 A0A1Q3FYB2 A0A1I8M3S5 A0A1I8NU89 A0A1W4XVD0 V5I8R8 U5EX18 E9J8K4 A0A158NBU8 K7ISE6 A0A2J7PX15 A0A1B6GJW3 A0A1B6IZT0 E2ACH8 A0A195BF10 A0A0C9QZS2 A0A067R983 A0A0A9Y0B4 A0A0C9R328 N6UAJ9 A0A069DWU5 A0A023F5S2 A0A224XE67 A0A0P4W4K3 A0A2P8ZJQ9 E0VK25 A0A0J7KLI9 A0A1B6DGS2 A0A0L7QWP9 A0A088AH86 A0A2G3AQQ1 F4X887 A0A195FDW1 V9I9B6 A0A2S2P3D1 A0A0N0BFV5 J9K6T5 A0A2A3ETU6 A0A195C9K4 A0A1B6BYD6 V9IAN3 A0A0P5P0E4 A0A0P6FYT9 A0A0P6EL28 A0A0P5MTA5 A0A0P6DXK2 A0A0P6G5U3 A0A0P5QZM6 A0A0P5RT68 A0A0P5K670 A0A0P5JVD7 A0A0P5RYG8 A0A0P5PUF0 A0A0N8C123 A0A0P5NT51 A0A0P6ET47 A0A0N8BXF5 A0A0P6DRX0 A0A0P5J4G0 A0A0P5KBT6 A0A151X9P3 A0A0P6G9T0 A0A0P5HDM1 A0A0P5S4Q2 A0A0P6ELN1 A0A0P5N8H0 A0A151J959 A0A0N8DUR1 A0A0N8C786 A0A0P6HE64 A0A3S3S074 A0A0K2UUH0 A0A132A7P0 A0A0P5BVX6
A0A0K8TY54 A0A034VKS9 B3NVT4 A0A1B0CED0 B4PZ07 A0A0A1WPY7 W8C5S6 B4JWU0 A0A1A9WDQ1 A0A1L8DVN8 B4NQ71 A0A0L0CKZ8 A0A0M4EWR3 B4L6V2 A0A1B0FI36 A0A1A9USV6 A0A1B0BB68 A0A1A9ZQI5 A0A1A9XCV0 A0A1Q3FY77 A0A1Y1LRA6 A0A1Q3FYB2 A0A1I8M3S5 A0A1I8NU89 A0A1W4XVD0 V5I8R8 U5EX18 E9J8K4 A0A158NBU8 K7ISE6 A0A2J7PX15 A0A1B6GJW3 A0A1B6IZT0 E2ACH8 A0A195BF10 A0A0C9QZS2 A0A067R983 A0A0A9Y0B4 A0A0C9R328 N6UAJ9 A0A069DWU5 A0A023F5S2 A0A224XE67 A0A0P4W4K3 A0A2P8ZJQ9 E0VK25 A0A0J7KLI9 A0A1B6DGS2 A0A0L7QWP9 A0A088AH86 A0A2G3AQQ1 F4X887 A0A195FDW1 V9I9B6 A0A2S2P3D1 A0A0N0BFV5 J9K6T5 A0A2A3ETU6 A0A195C9K4 A0A1B6BYD6 V9IAN3 A0A0P5P0E4 A0A0P6FYT9 A0A0P6EL28 A0A0P5MTA5 A0A0P6DXK2 A0A0P6G5U3 A0A0P5QZM6 A0A0P5RT68 A0A0P5K670 A0A0P5JVD7 A0A0P5RYG8 A0A0P5PUF0 A0A0N8C123 A0A0P5NT51 A0A0P6ET47 A0A0N8BXF5 A0A0P6DRX0 A0A0P5J4G0 A0A0P5KBT6 A0A151X9P3 A0A0P6G9T0 A0A0P5HDM1 A0A0P5S4Q2 A0A0P6ELN1 A0A0P5N8H0 A0A151J959 A0A0N8DUR1 A0A0N8C786 A0A0P6HE64 A0A3S3S074 A0A0K2UUH0 A0A132A7P0 A0A0P5BVX6
PDB
6DFF
E-value=0,
Score=2478
Ontologies
GO
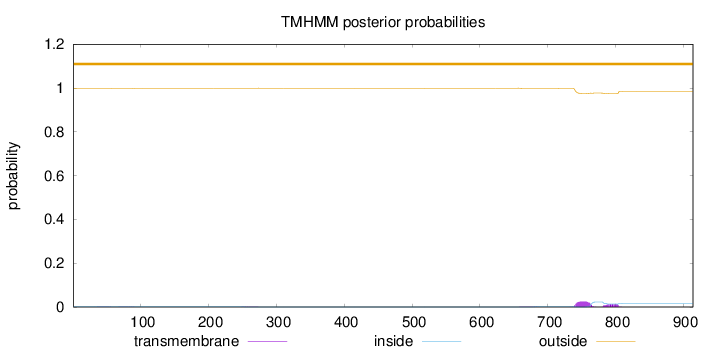
Topology
Subcellular location
Cytoplasm
Length:
914
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.858739999999998
Exp number, first 60 AAs:
0.0066
Total prob of N-in:
0.00200
outside
1 - 914
Population Genetic Test Statistics
Pi
244.867794
Theta
167.406256
Tajima's D
1.475001
CLR
0.068729
CSRT
0.780510974451277
Interpretation
Uncertain
