Gene
KWMTBOMO09775
Pre Gene Modal
BGIBMGA012915
Annotation
PREDICTED:_PAX-interacting_protein_1_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 2.382
Sequence
CDS
ATGCAGGCTCTCCGCGACGCGAAACGCTGCGTGACCGCGTACTGGCTGTCGGACGCGATGGTGCGGCGCGGCGTGGCCCCGCCGTGGCAGGCGCTGCACCTGCCGCTGCTGTACGCCGCGCGCGACCGCCCCGCGCGACACCACCGCGCCGCGCTCTCCGGCTGGCGGCGCGACGAGCGATCGCGACTCGTCTGCTGCCTCGAACACGTCGGGGCCAAGCTGACGCCGTACATGACGCGCGACAACACGGTGCTGGTGTGCAAGCGCGCCGAGGGCAACAAGTACCGGCGCGCCAAGGAGTGGGGCATCCCCGTGGTCACCGCCGCCTGGCTCACCGACCTGCTGCTCGGGAACATGACGGCGCTCGCGCAGATTGAAAATGCGAAATATCAACAATTTAACTTGGCGAGTCCGTTCCGAATGGATTACAGCTTGGTGTCGCACTTAATGAACGCGTGGAAGATGCCAATAAACATAACGCAAGAGTCGCACGAGCGGGCGAAGCGCGCGGCGGCGGCGGTGTGCGCGGGCGTGCGGCGCGCCAAGCGGCCCCGCCTCGAGTCCCCCCCGCCCCCCGCGCCGCACGCCGCGCACCCCCCGCTGCACCTCGCGCCGCGCGTGCTGTTCTCCGCCGTGCCGCCCGCCCTGCAGAACAAGCTCGCCACCGTCGTCAGGCAACTCGGAGGACTCGTCGTAACCTCAGTAGCCGAAGCGACGCACTTGGTAATGGAAAAGTTGGTGCGAACGTGCAAGCTGGTCAGCTGTCTCGTCACCGTCAAGCACTTGCTCACTCCCGAGTGGATCCTCGAGAGTCAAAGGCAGAATAAATTTGCGGACGAAGCGAAGCACGGTCTGCGGGACGAAGCCTTCAATAAAATGTTCCGTTGTGACGTGGCCGAGGTCCTGCTGTGCGGCGAGCAGAGGAAGAAGCTGTTTGAAGGCGTCACTTTCTTCATGACGCCGTGCGTGAAGCCAACAAAAGCCGCGCTCACCGAGATGATAGAGCTGTGCGGCGGCAGAGTCGAAAAGAGCCGCCGTTCATTTGTCTCCATACAGGAGATGCACTCGCAGAGACCGTACAGTTACTTGGTACTGACCGTGCCCAACGATCTCCATCTAGTGTATTATCTATTGCAGTCCGAGAAGACGTTGAACGTCGTTTGCAGTACCGAGGTTGTTCTGTCGGCGATCATGAGACAGAAGTTGGAGGTCGAAGAATTTCTCGTCAAAATCGACTAA
Protein
MQALRDAKRCVTAYWLSDAMVRRGVAPPWQALHLPLLYAARDRPARHHRAALSGWRRDERSRLVCCLEHVGAKLTPYMTRDNTVLVCKRAEGNKYRRAKEWGIPVVTAAWLTDLLLGNMTALAQIENAKYQQFNLASPFRMDYSLVSHLMNAWKMPINITQESHERAKRAAAAVCAGVRRAKRPRLESPPPPAPHAAHPPLHLAPRVLFSAVPPALQNKLATVVRQLGGLVVTSVAEATHLVMEKLVRTCKLVSCLVTVKHLLTPEWILESQRQNKFADEAKHGLRDEAFNKMFRCDVAEVLLCGEQRKKLFEGVTFFMTPCVKPTKAALTEMIELCGGRVEKSRRSFVSIQEMHSQRPYSYLVLTVPNDLHLVYYLLQSEKTLNVVCSTEVVLSAIMRQKLEVEEFLVKID
Summary
Uniprot
A0A194Q5Y2
A0A3S2P0Q4
A0A0N1IFY1
A0A212FMD8
A0A2A4J909
A0A1B0DJ43
+ More
A0A1B0CG64 A0A1Y1KSD2 A0A0L7QUT0 D2A4R5 A0A087ZPR4 A0A2A3EJX9 A0A182JVI3 A0A0C9QTH5 A0A0P6ITI3 A0A0M9A5Z4 A0A336K9F2 A0A347ZJ89 A0A2J7RMA5 A0A182U0M7 A0A182RZM0 A0A182QGW3 A0A182VZL4 A0A182Y3S3 A0A182NRU4 U5EWC0 B0XGQ5 A7UVI9 A0A182TXB5 E2AED1 A0A182M6X5 A0A182LA35 A0A182HLM9 K7J6R6 A0A182VID7 A0A182WSS6 A0A232EPX5 A0A0K8T797 A0A0A9X1P8 A0A182F7H3 A0A1W4WTP8 A0A2M4B936 A0A2M4A8I7 A0A2M4A4W6 A0A2M4CKW9 A0A2M4CL56 A0A158NXA2 A0A2M4CL84 A0A084VI15 A0A151JU79 A0A182PSJ5 A0A151WY89 A0A195BL42 B4HHB0 A0A034W7W4 F4WQ61 A0A026WD80 E9IVW4 A0A195ECQ5 A0A0A1X1D8 B4L165 B3MAG1 A0A151I7A8 A0A182IPY5 E2BRL1 B4H1W3 A0A034W683 A0A0K8UN87 B4LC32 A0A0J9RV86 A0A3B0JBP8 Q9VUB7 A0A310S856 B4MLV6 B3ND02 A0A023EZQ6 A0A1I8M668 A0A1W4UKA0 A0A1W4U7D9 A0A1I8PNI3 T1HG73 B4IXZ0 A0A0M5JAX5 A0A0L0BL10 A0A146M339 N6U322 B4QJJ4 A0A1J1HET3 A0A2R7VY81 A0A1A9WBC9 U4U5B3 A0A2M3YZ24 A0A0A9X764 A0A1B6MPS4 A0A2S2P967 A0A1B0GGK8
A0A1B0CG64 A0A1Y1KSD2 A0A0L7QUT0 D2A4R5 A0A087ZPR4 A0A2A3EJX9 A0A182JVI3 A0A0C9QTH5 A0A0P6ITI3 A0A0M9A5Z4 A0A336K9F2 A0A347ZJ89 A0A2J7RMA5 A0A182U0M7 A0A182RZM0 A0A182QGW3 A0A182VZL4 A0A182Y3S3 A0A182NRU4 U5EWC0 B0XGQ5 A7UVI9 A0A182TXB5 E2AED1 A0A182M6X5 A0A182LA35 A0A182HLM9 K7J6R6 A0A182VID7 A0A182WSS6 A0A232EPX5 A0A0K8T797 A0A0A9X1P8 A0A182F7H3 A0A1W4WTP8 A0A2M4B936 A0A2M4A8I7 A0A2M4A4W6 A0A2M4CKW9 A0A2M4CL56 A0A158NXA2 A0A2M4CL84 A0A084VI15 A0A151JU79 A0A182PSJ5 A0A151WY89 A0A195BL42 B4HHB0 A0A034W7W4 F4WQ61 A0A026WD80 E9IVW4 A0A195ECQ5 A0A0A1X1D8 B4L165 B3MAG1 A0A151I7A8 A0A182IPY5 E2BRL1 B4H1W3 A0A034W683 A0A0K8UN87 B4LC32 A0A0J9RV86 A0A3B0JBP8 Q9VUB7 A0A310S856 B4MLV6 B3ND02 A0A023EZQ6 A0A1I8M668 A0A1W4UKA0 A0A1W4U7D9 A0A1I8PNI3 T1HG73 B4IXZ0 A0A0M5JAX5 A0A0L0BL10 A0A146M339 N6U322 B4QJJ4 A0A1J1HET3 A0A2R7VY81 A0A1A9WBC9 U4U5B3 A0A2M3YZ24 A0A0A9X764 A0A1B6MPS4 A0A2S2P967 A0A1B0GGK8
Pubmed
26354079
22118469
28004739
18362917
19820115
26999592
+ More
25244985 12364791 14747013 17210077 20798317 20966253 20075255 28648823 25401762 21347285 24438588 17994087 25348373 21719571 24508170 30249741 21282665 25830018 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25474469 25315136 26108605 26823975 23537049
25244985 12364791 14747013 17210077 20798317 20966253 20075255 28648823 25401762 21347285 24438588 17994087 25348373 21719571 24508170 30249741 21282665 25830018 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25474469 25315136 26108605 26823975 23537049
EMBL
KQ459582
KPI98815.1
RSAL01000008
RVE54003.1
KQ460635
KPJ13436.1
+ More
AGBW02007651 OWR54896.1 NWSH01002600 PCG67883.1 AJVK01006197 AJVK01006198 AJWK01010786 GEZM01075177 JAV64224.1 KQ414731 KOC62370.1 KQ971344 EFA05726.1 KZ288227 PBC31784.1 GBYB01003962 JAG73729.1 GDUN01000887 JAN95032.1 KQ435746 KOX76549.1 UFQS01000118 UFQT01000118 SSX00005.1 SSX20385.1 FX985657 BBA84395.1 NEVH01002568 PNF41949.1 AXCN02000249 AXCN02000250 GANO01003018 JAB56853.1 DS233057 EDS27778.1 AAAB01008987 EDO63233.2 GL438827 EFN68364.1 AXCM01000026 APCN01000909 AAZX01000214 NNAY01002854 OXU20439.1 GBRD01004811 JAG61010.1 GBHO01028992 JAG14612.1 GGFJ01000423 MBW49564.1 GGFK01003607 MBW36928.1 GGFK01002523 MBW35844.1 GGFL01001643 MBW65821.1 GGFL01001787 MBW65965.1 ADTU01003030 GGFL01001918 MBW66096.1 ATLV01013256 ATLV01013257 ATLV01013258 KE524848 KFB37609.1 KQ981799 KYN35659.1 KQ982651 KYQ52862.1 KQ976441 KYM86387.1 CH480815 EDW41435.1 GAKP01008757 JAC50195.1 GL888262 EGI63684.1 KK107260 QOIP01000001 EZA54022.1 RLU26474.1 GL766449 EFZ15244.1 KQ979074 KYN22891.1 GBXI01009390 JAD04902.1 CH933809 EDW19247.2 CH902618 EDV40212.2 KQ978464 KYM93782.1 GL449965 EFN81703.1 CH479203 EDW30315.1 GAKP01008758 JAC50194.1 GDHF01024346 JAI27968.1 CH940647 EDW70860.2 CM002912 KMY99502.1 OUUW01000002 SPP77462.1 AE014296 AAF49771.3 KQ769958 OAD52708.1 CH963847 EDW73167.2 CH954178 EDV51724.2 GBBI01003965 JAC14747.1 ACPB03002065 CH916366 EDV97533.1 CP012525 ALC43481.1 JRES01001711 KNC20628.1 GDHC01004438 JAQ14191.1 APGK01044176 APGK01044177 KB741021 ENN75011.1 CM000363 EDX10403.1 CVRI01000001 CRK86347.1 KK854166 PTY12457.1 KB631749 ERL85786.1 GGFM01000755 MBW21506.1 GBHO01028991 JAG14613.1 GEBQ01002017 JAT37960.1 GGMR01013335 MBY25954.1 CCAG010023047
AGBW02007651 OWR54896.1 NWSH01002600 PCG67883.1 AJVK01006197 AJVK01006198 AJWK01010786 GEZM01075177 JAV64224.1 KQ414731 KOC62370.1 KQ971344 EFA05726.1 KZ288227 PBC31784.1 GBYB01003962 JAG73729.1 GDUN01000887 JAN95032.1 KQ435746 KOX76549.1 UFQS01000118 UFQT01000118 SSX00005.1 SSX20385.1 FX985657 BBA84395.1 NEVH01002568 PNF41949.1 AXCN02000249 AXCN02000250 GANO01003018 JAB56853.1 DS233057 EDS27778.1 AAAB01008987 EDO63233.2 GL438827 EFN68364.1 AXCM01000026 APCN01000909 AAZX01000214 NNAY01002854 OXU20439.1 GBRD01004811 JAG61010.1 GBHO01028992 JAG14612.1 GGFJ01000423 MBW49564.1 GGFK01003607 MBW36928.1 GGFK01002523 MBW35844.1 GGFL01001643 MBW65821.1 GGFL01001787 MBW65965.1 ADTU01003030 GGFL01001918 MBW66096.1 ATLV01013256 ATLV01013257 ATLV01013258 KE524848 KFB37609.1 KQ981799 KYN35659.1 KQ982651 KYQ52862.1 KQ976441 KYM86387.1 CH480815 EDW41435.1 GAKP01008757 JAC50195.1 GL888262 EGI63684.1 KK107260 QOIP01000001 EZA54022.1 RLU26474.1 GL766449 EFZ15244.1 KQ979074 KYN22891.1 GBXI01009390 JAD04902.1 CH933809 EDW19247.2 CH902618 EDV40212.2 KQ978464 KYM93782.1 GL449965 EFN81703.1 CH479203 EDW30315.1 GAKP01008758 JAC50194.1 GDHF01024346 JAI27968.1 CH940647 EDW70860.2 CM002912 KMY99502.1 OUUW01000002 SPP77462.1 AE014296 AAF49771.3 KQ769958 OAD52708.1 CH963847 EDW73167.2 CH954178 EDV51724.2 GBBI01003965 JAC14747.1 ACPB03002065 CH916366 EDV97533.1 CP012525 ALC43481.1 JRES01001711 KNC20628.1 GDHC01004438 JAQ14191.1 APGK01044176 APGK01044177 KB741021 ENN75011.1 CM000363 EDX10403.1 CVRI01000001 CRK86347.1 KK854166 PTY12457.1 KB631749 ERL85786.1 GGFM01000755 MBW21506.1 GBHO01028991 JAG14613.1 GEBQ01002017 JAT37960.1 GGMR01013335 MBY25954.1 CCAG010023047
Proteomes
UP000053268
UP000283053
UP000053240
UP000007151
UP000218220
UP000092462
+ More
UP000092461 UP000053825 UP000007266 UP000005203 UP000242457 UP000075881 UP000053105 UP000235965 UP000075902 UP000075900 UP000075886 UP000075920 UP000076408 UP000075884 UP000002320 UP000007062 UP000000311 UP000075883 UP000075882 UP000075840 UP000002358 UP000075903 UP000076407 UP000215335 UP000069272 UP000192223 UP000005205 UP000030765 UP000078541 UP000075885 UP000075809 UP000078540 UP000001292 UP000007755 UP000053097 UP000279307 UP000078492 UP000009192 UP000007801 UP000078542 UP000075880 UP000008237 UP000008744 UP000008792 UP000268350 UP000000803 UP000007798 UP000008711 UP000095301 UP000192221 UP000095300 UP000015103 UP000001070 UP000092553 UP000037069 UP000019118 UP000000304 UP000183832 UP000091820 UP000030742 UP000092444
UP000092461 UP000053825 UP000007266 UP000005203 UP000242457 UP000075881 UP000053105 UP000235965 UP000075902 UP000075900 UP000075886 UP000075920 UP000076408 UP000075884 UP000002320 UP000007062 UP000000311 UP000075883 UP000075882 UP000075840 UP000002358 UP000075903 UP000076407 UP000215335 UP000069272 UP000192223 UP000005205 UP000030765 UP000078541 UP000075885 UP000075809 UP000078540 UP000001292 UP000007755 UP000053097 UP000279307 UP000078492 UP000009192 UP000007801 UP000078542 UP000075880 UP000008237 UP000008744 UP000008792 UP000268350 UP000000803 UP000007798 UP000008711 UP000095301 UP000192221 UP000095300 UP000015103 UP000001070 UP000092553 UP000037069 UP000019118 UP000000304 UP000183832 UP000091820 UP000030742 UP000092444
PRIDE
SUPFAM
SSF52113
SSF52113
Gene 3D
CDD
ProteinModelPortal
A0A194Q5Y2
A0A3S2P0Q4
A0A0N1IFY1
A0A212FMD8
A0A2A4J909
A0A1B0DJ43
+ More
A0A1B0CG64 A0A1Y1KSD2 A0A0L7QUT0 D2A4R5 A0A087ZPR4 A0A2A3EJX9 A0A182JVI3 A0A0C9QTH5 A0A0P6ITI3 A0A0M9A5Z4 A0A336K9F2 A0A347ZJ89 A0A2J7RMA5 A0A182U0M7 A0A182RZM0 A0A182QGW3 A0A182VZL4 A0A182Y3S3 A0A182NRU4 U5EWC0 B0XGQ5 A7UVI9 A0A182TXB5 E2AED1 A0A182M6X5 A0A182LA35 A0A182HLM9 K7J6R6 A0A182VID7 A0A182WSS6 A0A232EPX5 A0A0K8T797 A0A0A9X1P8 A0A182F7H3 A0A1W4WTP8 A0A2M4B936 A0A2M4A8I7 A0A2M4A4W6 A0A2M4CKW9 A0A2M4CL56 A0A158NXA2 A0A2M4CL84 A0A084VI15 A0A151JU79 A0A182PSJ5 A0A151WY89 A0A195BL42 B4HHB0 A0A034W7W4 F4WQ61 A0A026WD80 E9IVW4 A0A195ECQ5 A0A0A1X1D8 B4L165 B3MAG1 A0A151I7A8 A0A182IPY5 E2BRL1 B4H1W3 A0A034W683 A0A0K8UN87 B4LC32 A0A0J9RV86 A0A3B0JBP8 Q9VUB7 A0A310S856 B4MLV6 B3ND02 A0A023EZQ6 A0A1I8M668 A0A1W4UKA0 A0A1W4U7D9 A0A1I8PNI3 T1HG73 B4IXZ0 A0A0M5JAX5 A0A0L0BL10 A0A146M339 N6U322 B4QJJ4 A0A1J1HET3 A0A2R7VY81 A0A1A9WBC9 U4U5B3 A0A2M3YZ24 A0A0A9X764 A0A1B6MPS4 A0A2S2P967 A0A1B0GGK8
A0A1B0CG64 A0A1Y1KSD2 A0A0L7QUT0 D2A4R5 A0A087ZPR4 A0A2A3EJX9 A0A182JVI3 A0A0C9QTH5 A0A0P6ITI3 A0A0M9A5Z4 A0A336K9F2 A0A347ZJ89 A0A2J7RMA5 A0A182U0M7 A0A182RZM0 A0A182QGW3 A0A182VZL4 A0A182Y3S3 A0A182NRU4 U5EWC0 B0XGQ5 A7UVI9 A0A182TXB5 E2AED1 A0A182M6X5 A0A182LA35 A0A182HLM9 K7J6R6 A0A182VID7 A0A182WSS6 A0A232EPX5 A0A0K8T797 A0A0A9X1P8 A0A182F7H3 A0A1W4WTP8 A0A2M4B936 A0A2M4A8I7 A0A2M4A4W6 A0A2M4CKW9 A0A2M4CL56 A0A158NXA2 A0A2M4CL84 A0A084VI15 A0A151JU79 A0A182PSJ5 A0A151WY89 A0A195BL42 B4HHB0 A0A034W7W4 F4WQ61 A0A026WD80 E9IVW4 A0A195ECQ5 A0A0A1X1D8 B4L165 B3MAG1 A0A151I7A8 A0A182IPY5 E2BRL1 B4H1W3 A0A034W683 A0A0K8UN87 B4LC32 A0A0J9RV86 A0A3B0JBP8 Q9VUB7 A0A310S856 B4MLV6 B3ND02 A0A023EZQ6 A0A1I8M668 A0A1W4UKA0 A0A1W4U7D9 A0A1I8PNI3 T1HG73 B4IXZ0 A0A0M5JAX5 A0A0L0BL10 A0A146M339 N6U322 B4QJJ4 A0A1J1HET3 A0A2R7VY81 A0A1A9WBC9 U4U5B3 A0A2M3YZ24 A0A0A9X764 A0A1B6MPS4 A0A2S2P967 A0A1B0GGK8
PDB
3SQD
E-value=9.06598e-20,
Score=239
Ontologies
Topology
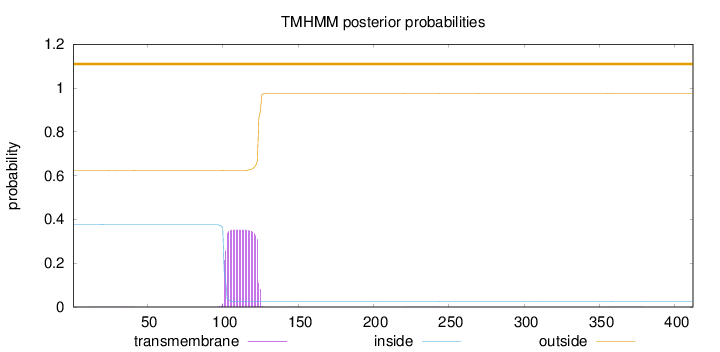
Length:
412
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.03031999999998
Exp number, first 60 AAs:
0.02397
Total prob of N-in:
0.37672
outside
1 - 412
Population Genetic Test Statistics
Pi
230.680752
Theta
159.865329
Tajima's D
1.616106
CLR
0.044848
CSRT
0.812359382030898
Interpretation
Uncertain
