Gene
KWMTBOMO09772
Pre Gene Modal
BGIBMGA012924
Annotation
PREDICTED:_N-glycosylase/DNA_lyase_isoform_X2_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.19 Nuclear Reliability : 2.022
Sequence
CDS
ATGGTGGAAAAGCTGTGCACAAATTATGGTGAAGAAATCTGTAAGCATGAAAATACTGTGCATTACTCCTTCCCTGATATTGAGAAATTGGAGTGCCCAAAAGTTGAAACACAACTAAGAAACCTAGGGTTTGGATACAGAGCGAAATTCATACAAAAATCAGCATCACAAATAGTTGAATGGGGTGGTGCGAAATGGTTTGATGGTCTTCAAAACATGAAGTACAAAGATGCTAAATGTGAACTCATGAAATTACACGGAATCGGACCTAAAGTCGCGGACTGCATTTGTCTTATGTCACTAAATCATCTCGAAGCACTGCCAGTGGACACGCATGTTTATCAGATAGCAGCCCAGAACTATTTACCACATTTACGAGGAAAGAAGAATGTAACAGAAAAGATGTACACAGAGATCGGAGATCATTTTAGAACGCTCTATGGTGATATGGCTGGGTGGGCTCACACTATTTTATTCTGCTCAGATTTAAAGAAATTCCAGCTTGGTGATAATTCTCAAGTAAATGAATGTTCAAAACCGAAAAGAAAAAGAAAAAAGTAA
Protein
MVEKLCTNYGEEICKHENTVHYSFPDIEKLECPKVETQLRNLGFGYRAKFIQKSASQIVEWGGAKWFDGLQNMKYKDAKCELMKLHGIGPKVADCICLMSLNHLEALPVDTHVYQIAAQNYLPHLRGKKNVTEKMYTEIGDHFRTLYGDMAGWAHTILFCSDLKKFQLGDNSQVNECSKPKRKRKK
Summary
Uniprot
H9JTR2
S4NSP3
A0A2A4JB61
A0A194PZR1
A0A3S2LHL9
A0A0N1IPC4
+ More
A0A212FMB8 A0A0L7KXR5 A0A2W1BUI9 A0A067QHL1 A0A1B0CTL2 A0A2J7QV26 B0WKR6 D6WPN3 A0A182VQU7 A0A1B6EGV1 A0A1W4WYJ9 E9G475 A0A1S4FIL4 A0A182MJ91 Q7PYB2 A0A182TW63 Q16JX8 A0A182XBQ1 A0A182I0I7 A0A182VAG4 N6TPV9 A0A182RBY0 A0A0N7ZBT1 A0A0P5GBS8 A0A0P4W234 A0A0N8E0F8 A0A0P5DEN6 A0A0P5L7B5 A0A182N4Y3 A0A0N7ZPB2 A0A182IST3 A0A084VRR6 A0A0N8D442 A0A2S2NPN2 A0A0P6CMU1 A0A0P5AYI1 A0A0P5SMX4 A0A0P5Z1A3 A0A0P6E540 A0A0P6CPD8 H2CP63 A0A182YJX6 A0A182T7M4 A0A2H8TVZ3 A0A182K2E5 A0A182F6X6 A0A0P6DQX9 A0A0P5H3P3 J9K746 A0A2S2RAM6 E0VWU9 A0A0P5NUT1 A0A0P6J3Y6 A0A182PKC7 A0A0K8T7Y0 A0A0A9W576 W5JJN5 A0A210R1C6 A0A182KM85 A0A0N8DYP6 A0A182QC32 A0A0A9YQW9 A0A224XCP5 A0A0P6GPD2 A0A1B6MEG0 A0A0P5A7A0 A0A0P5HB12 A0A162NVS3 A0A0P5WFE9 K1RSU3 A0A023F2P9 B4JX36 A0A087UL83 A0A1B6GIX5 U4U8S6 A0A1I8P114 A0A1B6FIX7 A0A3G1VUD0 A0A1B6L4H6 A0A077X2A0 A0A1A8EB80 Q16ZM0 A0A1A8Q9B9 A0A1A9VEL7 A0A0L0C421 V9LDS9 A7SGZ3 A0A1I8N3R6 A0A0J7KEW5 B4MA54 W5UE69 A0A401P0C1 A0A2C9K4U8 A0A2B4RZ77
A0A212FMB8 A0A0L7KXR5 A0A2W1BUI9 A0A067QHL1 A0A1B0CTL2 A0A2J7QV26 B0WKR6 D6WPN3 A0A182VQU7 A0A1B6EGV1 A0A1W4WYJ9 E9G475 A0A1S4FIL4 A0A182MJ91 Q7PYB2 A0A182TW63 Q16JX8 A0A182XBQ1 A0A182I0I7 A0A182VAG4 N6TPV9 A0A182RBY0 A0A0N7ZBT1 A0A0P5GBS8 A0A0P4W234 A0A0N8E0F8 A0A0P5DEN6 A0A0P5L7B5 A0A182N4Y3 A0A0N7ZPB2 A0A182IST3 A0A084VRR6 A0A0N8D442 A0A2S2NPN2 A0A0P6CMU1 A0A0P5AYI1 A0A0P5SMX4 A0A0P5Z1A3 A0A0P6E540 A0A0P6CPD8 H2CP63 A0A182YJX6 A0A182T7M4 A0A2H8TVZ3 A0A182K2E5 A0A182F6X6 A0A0P6DQX9 A0A0P5H3P3 J9K746 A0A2S2RAM6 E0VWU9 A0A0P5NUT1 A0A0P6J3Y6 A0A182PKC7 A0A0K8T7Y0 A0A0A9W576 W5JJN5 A0A210R1C6 A0A182KM85 A0A0N8DYP6 A0A182QC32 A0A0A9YQW9 A0A224XCP5 A0A0P6GPD2 A0A1B6MEG0 A0A0P5A7A0 A0A0P5HB12 A0A162NVS3 A0A0P5WFE9 K1RSU3 A0A023F2P9 B4JX36 A0A087UL83 A0A1B6GIX5 U4U8S6 A0A1I8P114 A0A1B6FIX7 A0A3G1VUD0 A0A1B6L4H6 A0A077X2A0 A0A1A8EB80 Q16ZM0 A0A1A8Q9B9 A0A1A9VEL7 A0A0L0C421 V9LDS9 A7SGZ3 A0A1I8N3R6 A0A0J7KEW5 B4MA54 W5UE69 A0A401P0C1 A0A2C9K4U8 A0A2B4RZ77
Pubmed
19121390
23622113
26354079
22118469
26227816
28756777
+ More
24845553 18362917 19820115 21292972 12364791 14747013 17210077 17510324 23537049 24438588 21983336 25244985 20566863 25401762 20920257 23761445 28812685 20966253 22992520 25474469 17994087 26108605 24402279 17615350 25315136 23127152 30297745 15562597
24845553 18362917 19820115 21292972 12364791 14747013 17210077 17510324 23537049 24438588 21983336 25244985 20566863 25401762 20920257 23761445 28812685 20966253 22992520 25474469 17994087 26108605 24402279 17615350 25315136 23127152 30297745 15562597
EMBL
BABH01003421
GAIX01013992
JAA78568.1
NWSH01002123
PCG69089.1
KQ459582
+ More
KPI98817.1 RSAL01000008 RVE54005.1 KQ460635 KPJ13434.1 AGBW02007651 OWR54898.1 JTDY01004729 KOB67824.1 KZ149898 PZC78659.1 KK853400 KDR07858.1 AJWK01027765 NEVH01010479 PNF32439.1 DS231975 EDS30008.1 KQ971354 EFA06198.1 GEDC01000177 JAS37121.1 GL732531 EFX85705.1 AXCM01003997 AAAB01008987 EAA01084.4 CH477987 EAT34590.1 APCN01002082 APGK01025891 KB740605 ENN80038.1 GDRN01078805 JAI62512.1 GDIQ01248445 JAK03280.1 GDRN01078806 JAI62511.1 GDIQ01074004 JAN20733.1 GDIP01158595 JAJ64807.1 GDIQ01173773 JAK77952.1 GDIP01225841 JAI97560.1 ATLV01015773 KE525036 KFB40660.1 GDIP01070711 JAM33004.1 GGMR01006516 MBY19135.1 GDIQ01090859 JAN03878.1 GDIP01192523 JAJ30879.1 GDIP01137740 JAL65974.1 GDIP01051594 JAM52121.1 GDIQ01082298 JAN12439.1 GDIP01011483 JAM92232.1 JN090125 AEM59540.1 GFXV01006658 MBW18463.1 GDIQ01074003 JAN20734.1 GDIQ01232543 GDIQ01076753 JAK19182.1 ABLF02037401 GGMS01017179 MBY86382.1 DS235824 EEB17855.1 GDIQ01137437 JAL14289.1 GDIQ01023563 JAN71174.1 GBRD01004177 GBRD01001421 JAG61644.1 GBHO01043584 GBHO01043583 GBHO01043582 GBHO01016662 GBHO01016661 GBHO01011680 GBHO01011678 JAG00020.1 JAG00021.1 JAG00022.1 JAG26942.1 JAG26943.1 JAG31924.1 JAG31926.1 ADMH02001329 ETN62979.1 NEDP02000938 OWF54701.1 GDIQ01078900 JAN15837.1 AXCN02000187 GBHO01011679 JAG31925.1 GFTR01007682 JAW08744.1 GDIQ01032680 GDIQ01032288 JAN62449.1 GEBQ01005702 JAT34275.1 GDIP01206638 JAJ16764.1 GDIQ01232544 JAK19181.1 LRGB01000531 KZS18307.1 GDIP01086907 JAM16808.1 JH817012 EKC37606.1 GBBI01003254 JAC15458.1 CH916376 EDV95312.1 KK120364 KFM78122.1 GECZ01007506 JAS62263.1 KB632095 ERL88758.1 GECZ01019618 JAS50151.1 MH143575 AYK27449.1 GEBQ01021411 JAT18566.1 LK023379 CDS13398.1 HAEA01014713 SBQ43193.1 CH477488 EAT40106.1 HAEG01011581 SBR90131.1 JRES01000940 KNC26986.1 JW878229 AFP10746.1 DS469656 EDO37002.1 LBMM01008506 KMQ88819.1 CH940655 EDW66113.1 JT413520 AHH40281.1 BFAA01001508 GCB66554.1 LSMT01000197 PFX23764.1
KPI98817.1 RSAL01000008 RVE54005.1 KQ460635 KPJ13434.1 AGBW02007651 OWR54898.1 JTDY01004729 KOB67824.1 KZ149898 PZC78659.1 KK853400 KDR07858.1 AJWK01027765 NEVH01010479 PNF32439.1 DS231975 EDS30008.1 KQ971354 EFA06198.1 GEDC01000177 JAS37121.1 GL732531 EFX85705.1 AXCM01003997 AAAB01008987 EAA01084.4 CH477987 EAT34590.1 APCN01002082 APGK01025891 KB740605 ENN80038.1 GDRN01078805 JAI62512.1 GDIQ01248445 JAK03280.1 GDRN01078806 JAI62511.1 GDIQ01074004 JAN20733.1 GDIP01158595 JAJ64807.1 GDIQ01173773 JAK77952.1 GDIP01225841 JAI97560.1 ATLV01015773 KE525036 KFB40660.1 GDIP01070711 JAM33004.1 GGMR01006516 MBY19135.1 GDIQ01090859 JAN03878.1 GDIP01192523 JAJ30879.1 GDIP01137740 JAL65974.1 GDIP01051594 JAM52121.1 GDIQ01082298 JAN12439.1 GDIP01011483 JAM92232.1 JN090125 AEM59540.1 GFXV01006658 MBW18463.1 GDIQ01074003 JAN20734.1 GDIQ01232543 GDIQ01076753 JAK19182.1 ABLF02037401 GGMS01017179 MBY86382.1 DS235824 EEB17855.1 GDIQ01137437 JAL14289.1 GDIQ01023563 JAN71174.1 GBRD01004177 GBRD01001421 JAG61644.1 GBHO01043584 GBHO01043583 GBHO01043582 GBHO01016662 GBHO01016661 GBHO01011680 GBHO01011678 JAG00020.1 JAG00021.1 JAG00022.1 JAG26942.1 JAG26943.1 JAG31924.1 JAG31926.1 ADMH02001329 ETN62979.1 NEDP02000938 OWF54701.1 GDIQ01078900 JAN15837.1 AXCN02000187 GBHO01011679 JAG31925.1 GFTR01007682 JAW08744.1 GDIQ01032680 GDIQ01032288 JAN62449.1 GEBQ01005702 JAT34275.1 GDIP01206638 JAJ16764.1 GDIQ01232544 JAK19181.1 LRGB01000531 KZS18307.1 GDIP01086907 JAM16808.1 JH817012 EKC37606.1 GBBI01003254 JAC15458.1 CH916376 EDV95312.1 KK120364 KFM78122.1 GECZ01007506 JAS62263.1 KB632095 ERL88758.1 GECZ01019618 JAS50151.1 MH143575 AYK27449.1 GEBQ01021411 JAT18566.1 LK023379 CDS13398.1 HAEA01014713 SBQ43193.1 CH477488 EAT40106.1 HAEG01011581 SBR90131.1 JRES01000940 KNC26986.1 JW878229 AFP10746.1 DS469656 EDO37002.1 LBMM01008506 KMQ88819.1 CH940655 EDW66113.1 JT413520 AHH40281.1 BFAA01001508 GCB66554.1 LSMT01000197 PFX23764.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000283053
UP000053240
UP000007151
+ More
UP000037510 UP000027135 UP000092461 UP000235965 UP000002320 UP000007266 UP000075920 UP000192223 UP000000305 UP000075883 UP000007062 UP000075902 UP000008820 UP000076407 UP000075840 UP000075903 UP000019118 UP000075900 UP000075884 UP000075880 UP000030765 UP000076408 UP000075901 UP000075881 UP000069272 UP000007819 UP000009046 UP000075885 UP000000673 UP000242188 UP000075882 UP000075886 UP000076858 UP000005408 UP000001070 UP000054359 UP000030742 UP000095300 UP000078200 UP000037069 UP000001593 UP000095301 UP000036403 UP000008792 UP000221080 UP000288216 UP000076420 UP000225706
UP000037510 UP000027135 UP000092461 UP000235965 UP000002320 UP000007266 UP000075920 UP000192223 UP000000305 UP000075883 UP000007062 UP000075902 UP000008820 UP000076407 UP000075840 UP000075903 UP000019118 UP000075900 UP000075884 UP000075880 UP000030765 UP000076408 UP000075901 UP000075881 UP000069272 UP000007819 UP000009046 UP000075885 UP000000673 UP000242188 UP000075882 UP000075886 UP000076858 UP000005408 UP000001070 UP000054359 UP000030742 UP000095300 UP000078200 UP000037069 UP000001593 UP000095301 UP000036403 UP000008792 UP000221080 UP000288216 UP000076420 UP000225706
PRIDE
Interpro
SUPFAM
SSF48150
SSF48150
Gene 3D
CDD
ProteinModelPortal
H9JTR2
S4NSP3
A0A2A4JB61
A0A194PZR1
A0A3S2LHL9
A0A0N1IPC4
+ More
A0A212FMB8 A0A0L7KXR5 A0A2W1BUI9 A0A067QHL1 A0A1B0CTL2 A0A2J7QV26 B0WKR6 D6WPN3 A0A182VQU7 A0A1B6EGV1 A0A1W4WYJ9 E9G475 A0A1S4FIL4 A0A182MJ91 Q7PYB2 A0A182TW63 Q16JX8 A0A182XBQ1 A0A182I0I7 A0A182VAG4 N6TPV9 A0A182RBY0 A0A0N7ZBT1 A0A0P5GBS8 A0A0P4W234 A0A0N8E0F8 A0A0P5DEN6 A0A0P5L7B5 A0A182N4Y3 A0A0N7ZPB2 A0A182IST3 A0A084VRR6 A0A0N8D442 A0A2S2NPN2 A0A0P6CMU1 A0A0P5AYI1 A0A0P5SMX4 A0A0P5Z1A3 A0A0P6E540 A0A0P6CPD8 H2CP63 A0A182YJX6 A0A182T7M4 A0A2H8TVZ3 A0A182K2E5 A0A182F6X6 A0A0P6DQX9 A0A0P5H3P3 J9K746 A0A2S2RAM6 E0VWU9 A0A0P5NUT1 A0A0P6J3Y6 A0A182PKC7 A0A0K8T7Y0 A0A0A9W576 W5JJN5 A0A210R1C6 A0A182KM85 A0A0N8DYP6 A0A182QC32 A0A0A9YQW9 A0A224XCP5 A0A0P6GPD2 A0A1B6MEG0 A0A0P5A7A0 A0A0P5HB12 A0A162NVS3 A0A0P5WFE9 K1RSU3 A0A023F2P9 B4JX36 A0A087UL83 A0A1B6GIX5 U4U8S6 A0A1I8P114 A0A1B6FIX7 A0A3G1VUD0 A0A1B6L4H6 A0A077X2A0 A0A1A8EB80 Q16ZM0 A0A1A8Q9B9 A0A1A9VEL7 A0A0L0C421 V9LDS9 A7SGZ3 A0A1I8N3R6 A0A0J7KEW5 B4MA54 W5UE69 A0A401P0C1 A0A2C9K4U8 A0A2B4RZ77
A0A212FMB8 A0A0L7KXR5 A0A2W1BUI9 A0A067QHL1 A0A1B0CTL2 A0A2J7QV26 B0WKR6 D6WPN3 A0A182VQU7 A0A1B6EGV1 A0A1W4WYJ9 E9G475 A0A1S4FIL4 A0A182MJ91 Q7PYB2 A0A182TW63 Q16JX8 A0A182XBQ1 A0A182I0I7 A0A182VAG4 N6TPV9 A0A182RBY0 A0A0N7ZBT1 A0A0P5GBS8 A0A0P4W234 A0A0N8E0F8 A0A0P5DEN6 A0A0P5L7B5 A0A182N4Y3 A0A0N7ZPB2 A0A182IST3 A0A084VRR6 A0A0N8D442 A0A2S2NPN2 A0A0P6CMU1 A0A0P5AYI1 A0A0P5SMX4 A0A0P5Z1A3 A0A0P6E540 A0A0P6CPD8 H2CP63 A0A182YJX6 A0A182T7M4 A0A2H8TVZ3 A0A182K2E5 A0A182F6X6 A0A0P6DQX9 A0A0P5H3P3 J9K746 A0A2S2RAM6 E0VWU9 A0A0P5NUT1 A0A0P6J3Y6 A0A182PKC7 A0A0K8T7Y0 A0A0A9W576 W5JJN5 A0A210R1C6 A0A182KM85 A0A0N8DYP6 A0A182QC32 A0A0A9YQW9 A0A224XCP5 A0A0P6GPD2 A0A1B6MEG0 A0A0P5A7A0 A0A0P5HB12 A0A162NVS3 A0A0P5WFE9 K1RSU3 A0A023F2P9 B4JX36 A0A087UL83 A0A1B6GIX5 U4U8S6 A0A1I8P114 A0A1B6FIX7 A0A3G1VUD0 A0A1B6L4H6 A0A077X2A0 A0A1A8EB80 Q16ZM0 A0A1A8Q9B9 A0A1A9VEL7 A0A0L0C421 V9LDS9 A7SGZ3 A0A1I8N3R6 A0A0J7KEW5 B4MA54 W5UE69 A0A401P0C1 A0A2C9K4U8 A0A2B4RZ77
PDB
1KO9
E-value=1.36378e-35,
Score=370
Ontologies
KEGG
GO
GO:0003684
GO:0006284
GO:0006289
GO:0008534
GO:0016829
GO:0003824
GO:0006285
GO:0034039
GO:0005634
GO:0006364
GO:0016021
GO:0140078
GO:0003677
GO:0007004
GO:0070987
GO:0005739
GO:0031518
GO:0007420
GO:0055013
GO:0043524
GO:0003313
GO:0010975
GO:0006281
GO:0004222
GO:0016638
GO:0005488
GO:0005515
GO:0009258
GO:0016155
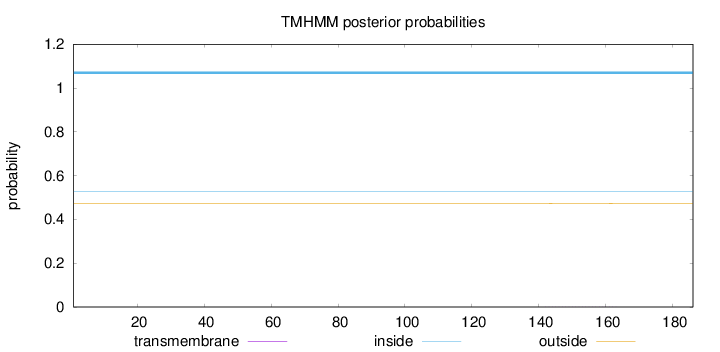
Topology
Length:
186
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01362
Exp number, first 60 AAs:
0.00011
Total prob of N-in:
0.52661
inside
1 - 186
Population Genetic Test Statistics
Pi
187.893035
Theta
144.24008
Tajima's D
1.162793
CLR
0.027293
CSRT
0.700314984250787
Interpretation
Uncertain
