Gene
KWMTBOMO09763
Pre Gene Modal
BGIBMGA012929
Annotation
PREDICTED:_phospholipase_A2-like_[Bombyx_mori]
Full name
Phospholipase A2
+ More
Phospholipase A(2)
Phospholipase A(2)
Alternative Name
Phosphatidylcholine 2-acylhydrolase
Allergen Api m I
Allergen Api m I
Location in the cell
Extracellular Reliability : 2.994
Sequence
CDS
ATGGCCGCTTGTTTATATTGTTTCTTTCTAATGATCGTTTTATACATTAACGGAGTGACCGGCAGCTGGTTCGCGAACGACGCTGCTGTTAATATCGATGAAGACGTACTAGATATTCAAAGTGAAACAATAGCAAGGAAGAAGTTCGACTTCATATTTCCTGGTACAAAATGGTGCGGTAAAGGGGACATAGCGAAGAACTACGAGGACCTGGGCTCAGCGCGCGAGACTGACATGTGCTGTCGGGAGCACGACCACTGCACTGACCTCATACTCGCCGGGGAGACCAAGCACGGCCTCACCAACGATGCCTTCTACACCAGGTTGAACTGTAAATGTGACGAGACATTCCGGCTGTGTCTGAACAAAGCTAACACGAAGACGTCGCGCAGAATCGGCAAGATATACTTCAACGCGCTCGGCACTAAGTGCTACAGACGAGACTACCCTGTCGTAGGCTGCAACACTTACGGAGGATTGTTCAGAAAGAAATGTTTGGAGTACGATTACGATACGAGGAGCGAACAGATCTACCAGTGGTTCGACGTCAGGAACTACGCCAACTGA
Protein
MAACLYCFFLMIVLYINGVTGSWFANDAAVNIDEDVLDIQSETIARKKFDFIFPGTKWCGKGDIAKNYEDLGSARETDMCCREHDHCTDLILAGETKHGLTNDAFYTRLNCKCDETFRLCLNKANTKTSRRIGKIYFNALGTKCYRRDYPVVGCNTYGGLFRKKCLEYDYDTRSEQIYQWFDVRNYAN
Summary
Description
PLA2 catalyzes the calcium-dependent hydrolysis of the 2-acyl groups in 3-sn-phosphoglycerides.
Catalytic Activity
a 1,2-diacyl-sn-glycero-3-phosphocholine + H2O = a 1-acyl-sn-glycero-3-phosphocholine + a fatty acid + H(+)
Cofactor
Ca(2+)
Miscellaneous
The secretion of this protein into venom follows a seasonal pattern. This variation is synchronized with melittin variation, i.e. their production increase in the same months.
Similarity
Belongs to the phospholipase A2 family.
Belongs to the phospholipase A2 family. Group III subfamily.
Belongs to the phospholipase A2 family. Group III subfamily.
Keywords
Allergen
Calcium
Direct protein sequencing
Disulfide bond
Glycoprotein
Hydrolase
Lipid degradation
Lipid metabolism
Metal-binding
Secreted
Signal
3D-structure
Complete proteome
Reference proteome
Feature
chain Phospholipase A2
glycosylation site N-linked (GlcNAc...) asparagine
glycosylation site N-linked (GlcNAc...) asparagine
Uniprot
H9JTR7
H9JTR6
H9JTQ0
H9JTP9
H9JTQ1
H9JTR4
+ More
A0A2H1VKV4 A0A437BU79 A0A3Q8U958 A0A212FMH8 A0A2A4ITF0 A0A2W1BZG9 U4V016 D6WFW7 J3JUT4 A0A1Y1LD35 A0A151IY19 A0A0C9QT97 A0A0T6B3S9 A0A154PF10 A0A2L2XXA1 A0A2L2YPZ8 A0A0L7R6K6 A0A151X980 A0A0M8ZRJ1 C7B2L3 A0A195CTK0 U3N068 G3FP81 E2BDL0 Q7M4I6 A0A2P6LH46 C0LTQ1 A0A087UYP2 A0A1W7RA22 A0A2P8YQI3 T1E1S9 A0A087UYP4 C0LTQ4 P82971 A0A087UYN9 B0W2R2 A0A182HBX1 Q7M4I5 Q16H86 A0A224XGJ6 A0A0C9S3A5 A0A218QXA5 A0A218QXE2 A0A2A3EI01 A0A224XFF0 W5JHV7 A0A2L2YIA1 A0A1J1J4Y0 A0A139WH39 A0A1J1HVT8 I7GQA7 A0A182WFX2 A0A2P6LAN4 A0A182SLS6 B7UUK1 P00630 A0A182FKB8 Q9BMK4 A0A182NC88 A0A084VSJ7 A0A170RBK2 A0A182PJB4 I1VC86 A0A182YKL8 A0A182RYX2 A0A336LWW5 Q7QGS9 A0A3L8DCJ6 A0A1S4H571 A0A182WYE6 A0A182UQ13 A0A182KVM6 A0A182I5H7 T1INX7 A0A182MAK3 A0A2D0PCW5 A0A170RBM5 A0A1W7RA16 A0A182QG10 A0A151I0C4 A0A2P6LH31 A0A139WH90 D6WLM2 A0A139WGU7 A0A1W4VE46 A0A2J7Q6T9 A0A336LI29 A0A0U1TZ64
A0A2H1VKV4 A0A437BU79 A0A3Q8U958 A0A212FMH8 A0A2A4ITF0 A0A2W1BZG9 U4V016 D6WFW7 J3JUT4 A0A1Y1LD35 A0A151IY19 A0A0C9QT97 A0A0T6B3S9 A0A154PF10 A0A2L2XXA1 A0A2L2YPZ8 A0A0L7R6K6 A0A151X980 A0A0M8ZRJ1 C7B2L3 A0A195CTK0 U3N068 G3FP81 E2BDL0 Q7M4I6 A0A2P6LH46 C0LTQ1 A0A087UYP2 A0A1W7RA22 A0A2P8YQI3 T1E1S9 A0A087UYP4 C0LTQ4 P82971 A0A087UYN9 B0W2R2 A0A182HBX1 Q7M4I5 Q16H86 A0A224XGJ6 A0A0C9S3A5 A0A218QXA5 A0A218QXE2 A0A2A3EI01 A0A224XFF0 W5JHV7 A0A2L2YIA1 A0A1J1J4Y0 A0A139WH39 A0A1J1HVT8 I7GQA7 A0A182WFX2 A0A2P6LAN4 A0A182SLS6 B7UUK1 P00630 A0A182FKB8 Q9BMK4 A0A182NC88 A0A084VSJ7 A0A170RBK2 A0A182PJB4 I1VC86 A0A182YKL8 A0A182RYX2 A0A336LWW5 Q7QGS9 A0A3L8DCJ6 A0A1S4H571 A0A182WYE6 A0A182UQ13 A0A182KVM6 A0A182I5H7 T1INX7 A0A182MAK3 A0A2D0PCW5 A0A170RBM5 A0A1W7RA16 A0A182QG10 A0A151I0C4 A0A2P6LH31 A0A139WH90 D6WLM2 A0A139WGU7 A0A1W4VE46 A0A2J7Q6T9 A0A336LI29 A0A0U1TZ64
EC Number
3.1.1.4
Pubmed
EMBL
BABH01003424
BABH01003423
ODYU01003111
SOQ41453.1
RSAL01000008
RVE54007.1
+ More
MH061374 AZL90156.1 AGBW02007651 OWR54900.1 NWSH01007341 PCG63001.1 KZ149898 PZC78657.1 KI209013 KI210158 KI210159 ERL95965.1 ERL96175.1 ERL96180.1 KQ971328 EEZ99784.2 BT126999 AEE61961.1 GEZM01059238 JAV71533.1 KQ980780 KYN13196.1 GBYB01003857 JAG73624.1 LJIG01009957 KRT82042.1 KQ434890 KZC10443.1 IAAA01000951 LAA00646.1 IAAA01047628 IAAA01047629 LAA10097.1 KQ414646 KOC66459.1 KQ982398 KYQ56870.1 KQ435889 KOX69497.1 FJ768907 FJ768908 ACU12492.1 KQ977279 KYN04023.1 KF214771 AGW23551.1 JF751030 AEO51763.1 GL447678 EFN86172.1 MWRG01000062 PRD37888.1 FJ768718 ACN42747.1 KK122319 KFM82481.1 GFAH01000398 JAV47991.1 PYGN01000431 PSN46508.1 GAKT01000015 JAA93047.1 KFM82483.1 FJ768721 ACN42750.1 KFM82478.1 DS231828 EDS29815.1 JXUM01032580 KQ560960 KXJ80209.1 CH478192 EAT33614.1 GFBG01000075 JAW07092.1 GBXR01000008 JAG85218.1 GEUW01000062 JAW06983.1 GEUW01000022 JAW07023.1 KZ288249 PBC31104.1 GFBG01000076 JAW07091.1 ADMH02001144 ETN63942.1 IAAA01020399 LAA07839.1 CVRI01000072 CRL07447.1 KQ971343 KYB27219.1 CVRI01000017 CRK90265.1 AB731659 BAM25049.1 MWRG01000625 PRD35635.1 FM203121 CAR56722.1 AF438408 EF373554 X16709 AF321087 ATLV01016045 KE525054 KFB40941.1 KU203673 ANB82417.1 JQ900380 AFI40558.1 UFQS01000176 UFQT01000176 SSX00773.1 SSX21153.1 AAAB01008823 EAA05503.4 QOIP01000010 RLU17599.1 APCN01003445 JH431228 AXCM01005795 HAHE01000501 SNX36715.1 KU203674 ANB82418.1 GFAH01000397 JAV47992.1 AXCN02001761 KQ976639 KYM78825.1 PRD37887.1 KYB27221.1 EFA03430.2 KYB27220.1 NEVH01017450 PNF24304.1 UFQS01004355 UFQT01004355 SSX16345.1 SSX35663.1 EU252426 ACD11980.1
MH061374 AZL90156.1 AGBW02007651 OWR54900.1 NWSH01007341 PCG63001.1 KZ149898 PZC78657.1 KI209013 KI210158 KI210159 ERL95965.1 ERL96175.1 ERL96180.1 KQ971328 EEZ99784.2 BT126999 AEE61961.1 GEZM01059238 JAV71533.1 KQ980780 KYN13196.1 GBYB01003857 JAG73624.1 LJIG01009957 KRT82042.1 KQ434890 KZC10443.1 IAAA01000951 LAA00646.1 IAAA01047628 IAAA01047629 LAA10097.1 KQ414646 KOC66459.1 KQ982398 KYQ56870.1 KQ435889 KOX69497.1 FJ768907 FJ768908 ACU12492.1 KQ977279 KYN04023.1 KF214771 AGW23551.1 JF751030 AEO51763.1 GL447678 EFN86172.1 MWRG01000062 PRD37888.1 FJ768718 ACN42747.1 KK122319 KFM82481.1 GFAH01000398 JAV47991.1 PYGN01000431 PSN46508.1 GAKT01000015 JAA93047.1 KFM82483.1 FJ768721 ACN42750.1 KFM82478.1 DS231828 EDS29815.1 JXUM01032580 KQ560960 KXJ80209.1 CH478192 EAT33614.1 GFBG01000075 JAW07092.1 GBXR01000008 JAG85218.1 GEUW01000062 JAW06983.1 GEUW01000022 JAW07023.1 KZ288249 PBC31104.1 GFBG01000076 JAW07091.1 ADMH02001144 ETN63942.1 IAAA01020399 LAA07839.1 CVRI01000072 CRL07447.1 KQ971343 KYB27219.1 CVRI01000017 CRK90265.1 AB731659 BAM25049.1 MWRG01000625 PRD35635.1 FM203121 CAR56722.1 AF438408 EF373554 X16709 AF321087 ATLV01016045 KE525054 KFB40941.1 KU203673 ANB82417.1 JQ900380 AFI40558.1 UFQS01000176 UFQT01000176 SSX00773.1 SSX21153.1 AAAB01008823 EAA05503.4 QOIP01000010 RLU17599.1 APCN01003445 JH431228 AXCM01005795 HAHE01000501 SNX36715.1 KU203674 ANB82418.1 GFAH01000397 JAV47992.1 AXCN02001761 KQ976639 KYM78825.1 PRD37887.1 KYB27221.1 EFA03430.2 KYB27220.1 NEVH01017450 PNF24304.1 UFQS01004355 UFQT01004355 SSX16345.1 SSX35663.1 EU252426 ACD11980.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000218220
UP000030742
UP000007266
+ More
UP000078492 UP000076502 UP000053825 UP000075809 UP000053105 UP000078542 UP000008237 UP000054359 UP000245037 UP000002320 UP000069940 UP000249989 UP000008820 UP000242457 UP000000673 UP000183832 UP000075920 UP000075901 UP000005203 UP000069272 UP000075884 UP000030765 UP000075885 UP000076408 UP000075900 UP000007062 UP000279307 UP000076407 UP000075903 UP000075882 UP000075840 UP000075883 UP000075886 UP000078540 UP000192221 UP000235965
UP000078492 UP000076502 UP000053825 UP000075809 UP000053105 UP000078542 UP000008237 UP000054359 UP000245037 UP000002320 UP000069940 UP000249989 UP000008820 UP000242457 UP000000673 UP000183832 UP000075920 UP000075901 UP000005203 UP000069272 UP000075884 UP000030765 UP000075885 UP000076408 UP000075900 UP000007062 UP000279307 UP000076407 UP000075903 UP000075882 UP000075840 UP000075883 UP000075886 UP000078540 UP000192221 UP000235965
Interpro
SUPFAM
SSF48619
SSF48619
Gene 3D
ProteinModelPortal
H9JTR7
H9JTR6
H9JTQ0
H9JTP9
H9JTQ1
H9JTR4
+ More
A0A2H1VKV4 A0A437BU79 A0A3Q8U958 A0A212FMH8 A0A2A4ITF0 A0A2W1BZG9 U4V016 D6WFW7 J3JUT4 A0A1Y1LD35 A0A151IY19 A0A0C9QT97 A0A0T6B3S9 A0A154PF10 A0A2L2XXA1 A0A2L2YPZ8 A0A0L7R6K6 A0A151X980 A0A0M8ZRJ1 C7B2L3 A0A195CTK0 U3N068 G3FP81 E2BDL0 Q7M4I6 A0A2P6LH46 C0LTQ1 A0A087UYP2 A0A1W7RA22 A0A2P8YQI3 T1E1S9 A0A087UYP4 C0LTQ4 P82971 A0A087UYN9 B0W2R2 A0A182HBX1 Q7M4I5 Q16H86 A0A224XGJ6 A0A0C9S3A5 A0A218QXA5 A0A218QXE2 A0A2A3EI01 A0A224XFF0 W5JHV7 A0A2L2YIA1 A0A1J1J4Y0 A0A139WH39 A0A1J1HVT8 I7GQA7 A0A182WFX2 A0A2P6LAN4 A0A182SLS6 B7UUK1 P00630 A0A182FKB8 Q9BMK4 A0A182NC88 A0A084VSJ7 A0A170RBK2 A0A182PJB4 I1VC86 A0A182YKL8 A0A182RYX2 A0A336LWW5 Q7QGS9 A0A3L8DCJ6 A0A1S4H571 A0A182WYE6 A0A182UQ13 A0A182KVM6 A0A182I5H7 T1INX7 A0A182MAK3 A0A2D0PCW5 A0A170RBM5 A0A1W7RA16 A0A182QG10 A0A151I0C4 A0A2P6LH31 A0A139WH90 D6WLM2 A0A139WGU7 A0A1W4VE46 A0A2J7Q6T9 A0A336LI29 A0A0U1TZ64
A0A2H1VKV4 A0A437BU79 A0A3Q8U958 A0A212FMH8 A0A2A4ITF0 A0A2W1BZG9 U4V016 D6WFW7 J3JUT4 A0A1Y1LD35 A0A151IY19 A0A0C9QT97 A0A0T6B3S9 A0A154PF10 A0A2L2XXA1 A0A2L2YPZ8 A0A0L7R6K6 A0A151X980 A0A0M8ZRJ1 C7B2L3 A0A195CTK0 U3N068 G3FP81 E2BDL0 Q7M4I6 A0A2P6LH46 C0LTQ1 A0A087UYP2 A0A1W7RA22 A0A2P8YQI3 T1E1S9 A0A087UYP4 C0LTQ4 P82971 A0A087UYN9 B0W2R2 A0A182HBX1 Q7M4I5 Q16H86 A0A224XGJ6 A0A0C9S3A5 A0A218QXA5 A0A218QXE2 A0A2A3EI01 A0A224XFF0 W5JHV7 A0A2L2YIA1 A0A1J1J4Y0 A0A139WH39 A0A1J1HVT8 I7GQA7 A0A182WFX2 A0A2P6LAN4 A0A182SLS6 B7UUK1 P00630 A0A182FKB8 Q9BMK4 A0A182NC88 A0A084VSJ7 A0A170RBK2 A0A182PJB4 I1VC86 A0A182YKL8 A0A182RYX2 A0A336LWW5 Q7QGS9 A0A3L8DCJ6 A0A1S4H571 A0A182WYE6 A0A182UQ13 A0A182KVM6 A0A182I5H7 T1INX7 A0A182MAK3 A0A2D0PCW5 A0A170RBM5 A0A1W7RA16 A0A182QG10 A0A151I0C4 A0A2P6LH31 A0A139WH90 D6WLM2 A0A139WGU7 A0A1W4VE46 A0A2J7Q6T9 A0A336LI29 A0A0U1TZ64
PDB
1POC
E-value=1.07e-32,
Score=346
Ontologies
PATHWAY
00564
Glycerophospholipid metabolism - Bombyx mori (domestic silkworm)
00565 Ether lipid metabolism - Bombyx mori (domestic silkworm)
00590 Arachidonic acid metabolism - Bombyx mori (domestic silkworm)
00591 Linoleic acid metabolism - Bombyx mori (domestic silkworm)
00592 alpha-Linolenic acid metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00565 Ether lipid metabolism - Bombyx mori (domestic silkworm)
00590 Arachidonic acid metabolism - Bombyx mori (domestic silkworm)
00591 Linoleic acid metabolism - Bombyx mori (domestic silkworm)
00592 alpha-Linolenic acid metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
PANTHER
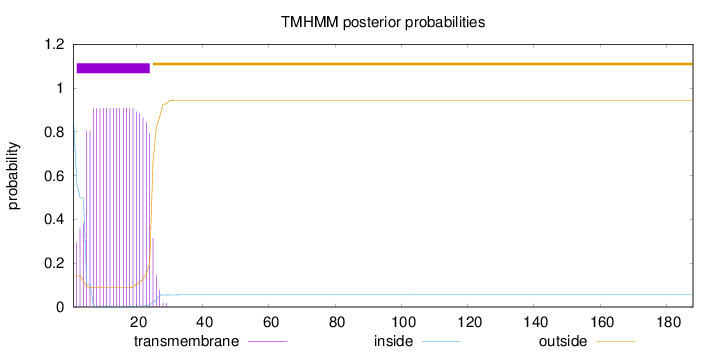
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 21,
Likelihood: 0.969767

Length:
188
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.30389
Exp number, first 60 AAs:
19.30276
Total prob of N-in:
0.85853
POSSIBLE N-term signal
sequence
inside
1 - 1
TMhelix
2 - 24
outside
25 - 188
Population Genetic Test Statistics
Pi
386.863414
Theta
212.654768
Tajima's D
2.647558
CLR
0
CSRT
0.955102244887756
Interpretation
Uncertain
