Gene
KWMTBOMO09761
Pre Gene Modal
BGIBMGA012908
Annotation
PREDICTED:_peripheral-type_benzodiazepine_receptor-associated_protein_1_isoform_X5_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.52
Sequence
CDS
ATGGATGTTCGAAATTCACTTTACATGAACGGGGCATCCGCTGAAACGGTGGACATGTTGGATATTCCCGGACGAGGCCGTTGTTGCGTTTACATAGCACGTTTCTCCTACGACCCTCCGGAAATCGAAAGCGCAGAGGGCGAATTATCGATTTCGGCGGGTGATTATTTATTAGTATGGGGCGCGCCGGATCCAAATGTAACAATGTTGGAAGCGGAAATGTTAGACGGGCGTCGCGGTCTTGTGCCAGTTAATTTTGTTCAGAGACTAGTGGGAGAGGATCTTTTAGACTTCCATCAAGCTGTGGTGACGTCGATGCGAGATGTTGATGAGTCTGCGACGACTGCGGTCAGTGAATTGTCTCTTAGTAGGGACGTCGCTCGACTTAGTGAAACGGCGGACATTAATGAGGGTCCTGAAGATGACGACAATGTGCCCGCGCCGCGGCAGCTCAACCTCGAGAGGCAATTGAATAAGTCCGTGCTGATCAGCTGGATCGCGCCGGACGGAGTTCAACAGGCGAACATCGAGAGCTATCACGTGTATGTCGATGGAGTCCTCAAAACTACTGTGAAAGCTACAGAACGCACGAGGGCCCTTGTCGAAGGAGTCGATGCTGGTCGCCCTCATAGAATAAGCGTGCGTTCCGTAACTGCTTCACGGAGGACATCACGTGATGCTGCTTGCACGATGGTCATAGGAAAGGAGACTGTTAACATGGGCCCAACTTGTGTCAGGGCTAGTGGAGTGACTTGTTCTCAAGCCGTCATTTCCTGGTTGCCTGCCAATTCTAATCAGCAGCACATAGTTTGCGTTAACAATGTTGAGGTGCGGCTAGTGAAAGCAGGTGTCTATCGGCATACCATTACGGGACTTTCACCCAGTACTCAGTACAGAGTAACAGTACGGGCCAAAAATGGGAAGACTCTCCAGCCTTCGGGAATACCCGCAGAAGAAGCTCCAGGCGCTTACACGGACTTTAGGACTCTACCCAAAGGCTTACCGGATCCGCCCAATGAAATAATGGTGGAGGCCGGCCCGCAAGACGGCACTCTGCTGGTGACGTGGCAGCCGGTACAGCGTCCGCCCGCCTCTGGACCTGTAACCGGATACGCTGTATACGCTGACGGAAAAAAGGTCACGGACGTAGACTCGCCGACCGGTGACCACGCTCTCATCGATATCGGCAAACTGATCGGCCTCAATCCCAAATGTGTTACCGTTCGTACAAAATCGCGCGACAGCCAATCCAGTGATAGTGTACCGACGCCTATTCCGCCTGCGGTATTACGGGGCGTCGTATCGAGAATGCCTCGTGGTGCAGTCGCGCCGGTCTCGATTCAGCCGGGCACGTACAGGGGCCCTCAGCGACCGCAGCCCTATCAGCAACAGCAACAAGTCATTGAGCATGACGAAAATTTATCAGATAAGGAAATATTCCCAAACAATAACCGCCACGATCAGCAGCCAGCCCAGCAACCGTCCCAACCGGCGCAGGCGTCGCAACCGCCTTCGGGTCAGCAGTACCCGAATACACCAGCGTACCATGGCACGCAGCAACCGCCGTATCCTCAAGTCGCCCAGCTCCCAAGTAACCAGCAGGGTCAGTACCCTAACCAGCCGCAGCAGTACCAGAATCCACCACCAAGGACACACCACGAAAGAGGACGACCACCGAATCCTCAGCAGCACGGGCAGCCAAGCGGGATGCAATTGCAAGGACGAGGGGCAGCGGCTGCGGGTGGCGCGGTACCTCGTGGCCCGCAACCGGGTCAACGTCATGGACCCCAGAGCCACGCTCAATCCAAACGCAGCCGCTTCTTCATCGCGCTCTTTGATTACGATCCAGTTACAATGAGTCCGAATCCAGAGAGTTGTGACGAAGAATTGCCCTTCAGCGAGGGTGACACAATAAAGGTATGGGGGGACAAAGACGCTGACGGTTTCTACTGGGGCGAGTGCCGAGGCCGACGCGGATATGTGCCGCACAACATGGTTATGGAAGTTTCCGAACAAGAGGCCACGGGGCAACCTGCCGCCAAGCCAAGAGACCGATGGACCGACGCCTATGCTAATCAACCCGTTCGAAGAATGGTCGCTCTCTATGACTATGATCCGCAAGAGCTGAGCCCCAATGTAGATGCCGATGCGGAATTAAGTTTCCAGACTGGTCAAATTATCCACGTTTACGGTGATATGGATGATGACGGTTTCTACATGGCTGAAATCGACGGAGTCCGAGGACTGGTCCCCAGCAATTTCCTCACTGACGCCAACGAACAGTATACCAATGCCGGACAGAGTAGTCAAGTTGGCGGCCCAAATATGCGCGGATCGGGCGGGGTCGCAGGGCGCGGTAGGGGCATGGGCCCGGGGGCGTCCGGGGGGCCAGGGGGCTCTGGGCCGCCGGGACCGGGGGCGCGCGGGCCCCCTCCCCCTCCGAGAGACAATGTTGCCCCCGCGCCCCGCAACCATCACCGTAAGACAGATGCCTGCCCTCTTCCCCCTTCTCAATTAGACCACAACACATGCGCCAGTAATCCAGAACAGGCGAATTCTCAGGCGAGGGGGAGAGGCGCGTCGAGCGGACAGCCCCCGAACACGATCGCGCGAACGAGCGCTGGTAATGCGGGCACCCCCACCGTCCAGTCGCCCGGCAACGCACCCCCCAGCGGACCCCCCACCTCCAACAACTCCGCGAGCACTCCGCTTAATGCGACCGGGCCCACTAGCACAACGACGGGGGGTCCTACGGCGCAAACAACTACCAGACGCGGAGGTCCAGCGGTCGTGCCTTCCGCGCAGCCCCTCACGCCGGCGCAGCCCCCCTCATCCCAGCCCAACCTGATGCAGAAGTTCTCCGAAATGACGGCACCCGGTGGCGACATTCTTAGTAAAGGCAAAGAACTGATCTTCATGAAATTCGGTCTCGGTGGTAAATAA
Protein
MDVRNSLYMNGASAETVDMLDIPGRGRCCVYIARFSYDPPEIESAEGELSISAGDYLLVWGAPDPNVTMLEAEMLDGRRGLVPVNFVQRLVGEDLLDFHQAVVTSMRDVDESATTAVSELSLSRDVARLSETADINEGPEDDDNVPAPRQLNLERQLNKSVLISWIAPDGVQQANIESYHVYVDGVLKTTVKATERTRALVEGVDAGRPHRISVRSVTASRRTSRDAACTMVIGKETVNMGPTCVRASGVTCSQAVISWLPANSNQQHIVCVNNVEVRLVKAGVYRHTITGLSPSTQYRVTVRAKNGKTLQPSGIPAEEAPGAYTDFRTLPKGLPDPPNEIMVEAGPQDGTLLVTWQPVQRPPASGPVTGYAVYADGKKVTDVDSPTGDHALIDIGKLIGLNPKCVTVRTKSRDSQSSDSVPTPIPPAVLRGVVSRMPRGAVAPVSIQPGTYRGPQRPQPYQQQQQVIEHDENLSDKEIFPNNNRHDQQPAQQPSQPAQASQPPSGQQYPNTPAYHGTQQPPYPQVAQLPSNQQGQYPNQPQQYQNPPPRTHHERGRPPNPQQHGQPSGMQLQGRGAAAAGGAVPRGPQPGQRHGPQSHAQSKRSRFFIALFDYDPVTMSPNPESCDEELPFSEGDTIKVWGDKDADGFYWGECRGRRGYVPHNMVMEVSEQEATGQPAAKPRDRWTDAYANQPVRRMVALYDYDPQELSPNVDADAELSFQTGQIIHVYGDMDDDGFYMAEIDGVRGLVPSNFLTDANEQYTNAGQSSQVGGPNMRGSGGVAGRGRGMGPGASGGPGGSGPPGPGARGPPPPPRDNVAPAPRNHHRKTDACPLPPSQLDHNTCASNPEQANSQARGRGASSGQPPNTIARTSAGNAGTPTVQSPGNAPPSGPPTSNNSASTPLNATGPTSTTTGGPTAQTTTRRGGPAVVPSAQPLTPAQPPSSQPNLMQKFSEMTAPGGDILSKGKELIFMKFGLGGK
Summary
Uniprot
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
PDB
4Z89
E-value=5.43691e-27,
Score=305
Ontologies
GO
PANTHER
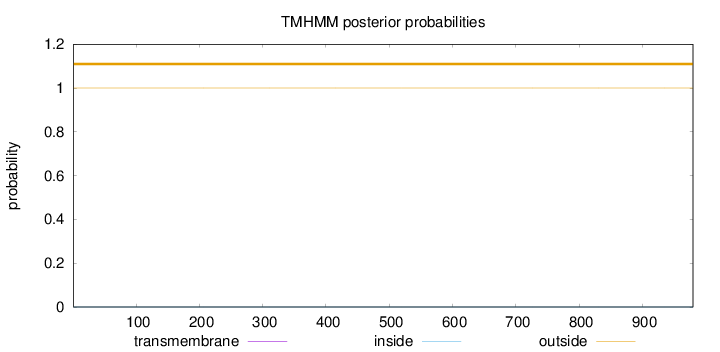
Topology
Length:
980
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000600000000000001
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.00005
outside
1 - 980
Population Genetic Test Statistics
Pi
279.307993
Theta
205.486337
Tajima's D
0.781535
CLR
0
CSRT
0.599470026498675
Interpretation
Uncertain
