Gene
KWMTBOMO09759
Pre Gene Modal
BGIBMGA012907
Annotation
CRAL-TRIO_domain-containing_protein?_partial_[Manduca_sexta]
Location in the cell
Cytoplasmic Reliability : 1.918 Nuclear Reliability : 1.353
Sequence
CDS
ATGGCTGCCACTAAATATCTGCACCCGTTTCTCAACGGCTTGAAGCTTTTGCCTGAAGGACATGAGGATGAAGTTCAGCAGATAAGAGAATGGATGAAAAGTCAACCCCACCTGCCGTATATATCGGACGAATACGTATATTTGTTTCTTCACTCAAACTATTACAAAGTGAAAGAGACGAAAGACACAATTGAATGCTACTTCACACTTCGTGGCAACACCCCTGATCTCTTTACGAACAGAGATCCTTTGGCTCCCAAGAATAGAGCTGTTATGGATATTACACAAATGGTAGCACTCCCAAATAAGACAACTGAAGGATATCATGTTCTTCTGTACCGCCTTGCCGACTATGATTATAGTAAGCTAAACTTCGCCGATGCTGTTCGCGTCTTTTGCATGTTCAATGATGTCAAACTCTCAGAGGATAAGCTCTCTGAAGGATACATCGTCATCTTTGACATGAAGGGATGTAGTCTCGGACATTTGACCAGGGTGACCCTGCCGGCGTTGAGAGCTTTCATGCACTACATCCAGAATGCTCATCCGTGTCGTCTAAAGAAGATCCACGTTGTTCACACGGTTTCCTTCATCAACCAGGTCATGTGTCTCGTGAAGCCACTCATACACTCCAATCTGTTGAACCTACTGAATTTCTCTTCGGAAGGCCCCGAATCAGTGGTCGATAAGCAACTCCTACCCGAAGAATTCGGTGGACCGATCGTTCCTGTGAAACAGCTACACGACGAACAGAGGAAGAACATGGAGGAGAACTACAGAGATTGGCTAATAGAAACGGAAATATTTAAAGCGGATGAGAAGAAGAGAATAAAGAAACCAAGTAAAGGGATGTTCGCAAGTTTCACGAGTAGCTTCAAAAGCTTAGAAATAGATTAA
Protein
MAATKYLHPFLNGLKLLPEGHEDEVQQIREWMKSQPHLPYISDEYVYLFLHSNYYKVKETKDTIECYFTLRGNTPDLFTNRDPLAPKNRAVMDITQMVALPNKTTEGYHVLLYRLADYDYSKLNFADAVRVFCMFNDVKLSEDKLSEGYIVIFDMKGCSLGHLTRVTLPALRAFMHYIQNAHPCRLKKIHVVHTVSFINQVMCLVKPLIHSNLLNLLNFSSEGPESVVDKQLLPEEFGGPIVPVKQLHDEQRKNMEENYRDWLIETEIFKADEKKRIKKPSKGMFASFTSSFKSLEID
Summary
Uniprot
H9JTP5
A0A0S2Z3D7
A0A2W1C113
A0A0L7LMY9
A0A437BV40
A0A212FMC8
+ More
A0A194R6X1 A0A2H4RMP9 A0A194Q1G6 A0A2H4RMW5 A0A2H1V5I9 A0A2A4JGQ4 J3JXS5 A0A182QUC4 A0A182VY11 A0A182IJU1 A0A182R5Q8 W5JB24 A0A182YP37 A0A182LYI0 A0A182TXX4 A0A182WVC3 A0A182LAZ2 Q7QG15 A0A182I904 A0A2M4AYU9 A0A182PKF8 A0A182N4K3 A0A2M4BWZ8 A0A182F947 A0A2M4BW08 A0A182UZY6 Q16RH7 A0A084VZX4 A0A182K9E8 E0W2C7 A0A023ENV8 A0A182H7H8 A0A336K9A5 A0A2M4BUS0 A0A1L8DBN2 A0A1L8DAM2 A0A2P8XI41 A0A1B6KRS7 A0A1B6LW17 A0A1B0CL60 A0A1J1IP62 A0A023F6J1 A0A069DR11 A0A224XH32 A0A0V0G5X2 B0W3K3 A0A1B6DLR3 E0W2C8 A0A1B6LD20 A0A1B6KYX9 A0A1W4X5F9 A0A1Y1JXE4 A0A1B6DYJ3 A0A0P4VRB9 R4G5W8 D6W9C3 A0A1Q3FQZ8 A0A1B6MQ64 A0A1B6DDH3 A0A1B6MKT4 A0A1Y1LMD7 A0A2S2Q3J3 A0A224XHT0 A0A1B6JB60 A0A2J7PS36 A0A023F9V9 A0A154PLP4 A0A0S2Z365 A0A1B6ISW4 T1HQM6 A0A1W4VFQ6 K7ITY7 A0A2S2N8X6 A0A310SB26 A0A0J7NH54 A0A0L0CKW7 T1HWT6 A0A2H8TII4 A0A067R4X7 E2BL70 V9IMD6 A0A151IIH9 A0A1L8DZD6 A0A151WMV2 K7J0H5 A0A0J7KK55 A0A0L7R9L5 A0A0L7R9P5 E2A8R0 B3NXR4 A0A2H4RMS2
A0A194R6X1 A0A2H4RMP9 A0A194Q1G6 A0A2H4RMW5 A0A2H1V5I9 A0A2A4JGQ4 J3JXS5 A0A182QUC4 A0A182VY11 A0A182IJU1 A0A182R5Q8 W5JB24 A0A182YP37 A0A182LYI0 A0A182TXX4 A0A182WVC3 A0A182LAZ2 Q7QG15 A0A182I904 A0A2M4AYU9 A0A182PKF8 A0A182N4K3 A0A2M4BWZ8 A0A182F947 A0A2M4BW08 A0A182UZY6 Q16RH7 A0A084VZX4 A0A182K9E8 E0W2C7 A0A023ENV8 A0A182H7H8 A0A336K9A5 A0A2M4BUS0 A0A1L8DBN2 A0A1L8DAM2 A0A2P8XI41 A0A1B6KRS7 A0A1B6LW17 A0A1B0CL60 A0A1J1IP62 A0A023F6J1 A0A069DR11 A0A224XH32 A0A0V0G5X2 B0W3K3 A0A1B6DLR3 E0W2C8 A0A1B6LD20 A0A1B6KYX9 A0A1W4X5F9 A0A1Y1JXE4 A0A1B6DYJ3 A0A0P4VRB9 R4G5W8 D6W9C3 A0A1Q3FQZ8 A0A1B6MQ64 A0A1B6DDH3 A0A1B6MKT4 A0A1Y1LMD7 A0A2S2Q3J3 A0A224XHT0 A0A1B6JB60 A0A2J7PS36 A0A023F9V9 A0A154PLP4 A0A0S2Z365 A0A1B6ISW4 T1HQM6 A0A1W4VFQ6 K7ITY7 A0A2S2N8X6 A0A310SB26 A0A0J7NH54 A0A0L0CKW7 T1HWT6 A0A2H8TII4 A0A067R4X7 E2BL70 V9IMD6 A0A151IIH9 A0A1L8DZD6 A0A151WMV2 K7J0H5 A0A0J7KK55 A0A0L7R9L5 A0A0L7R9P5 E2A8R0 B3NXR4 A0A2H4RMS2
Pubmed
EMBL
BABH01003428
KT943549
ALQ33307.1
KZ149898
PZC78656.1
JTDY01000521
+ More
KOB76812.1 RSAL01000008 RVE54009.1 AGBW02007651 OWR54902.1 KQ460878 KPJ11611.1 MG434602 ATY51908.1 KQ459582 KPI98834.1 MG434666 ATY51972.1 ODYU01000795 SOQ36115.1 NWSH01001518 PCG70999.1 BT128046 AEE63007.1 AXCN02000044 ADMH02001893 ETN60608.1 AXCM01013815 AAAB01008844 EAA06063.5 APCN01003147 GGFK01012571 MBW45892.1 GGFJ01008107 MBW57248.1 GGFJ01008108 MBW57249.1 CH477709 EAT37012.1 ATLV01019017 ATLV01019018 KE525259 KFB43518.1 DS235877 EEB19783.1 GAPW01002962 JAC10636.1 JXUM01028264 JXUM01030655 KQ560891 KQ560816 KXJ80465.1 KXJ80771.1 UFQS01000022 UFQT01000022 SSW97610.1 SSX17996.1 GGFJ01007550 MBW56691.1 GFDF01010299 JAV03785.1 GFDF01010667 JAV03417.1 PYGN01002057 PSN31652.1 GEBQ01025852 JAT14125.1 GEBQ01012094 JAT27883.1 AJWK01016944 AJWK01016945 AJWK01016946 AJWK01016947 AJWK01016948 AJWK01016949 CVRI01000057 CRL02023.1 GBBI01001854 JAC16858.1 GBGD01002381 JAC86508.1 GFTR01004631 JAW11795.1 GECL01003431 JAP02693.1 DS231832 EDS31789.1 GEDC01022556 GEDC01010710 GEDC01003689 GEDC01002746 JAS14742.1 JAS26588.1 JAS33609.1 JAS34552.1 EEB19784.1 GEBQ01018603 JAT21374.1 GEBQ01023362 JAT16615.1 GEZM01098136 JAV53982.1 GEDC01006573 JAS30725.1 GDKW01002174 JAI54421.1 GAHY01000155 JAA77355.1 KQ971312 EEZ98182.1 GFDL01005015 JAV30030.1 GEBQ01001890 JAT38087.1 GEDC01013602 JAS23696.1 GEBQ01003498 JAT36479.1 GEZM01051567 JAV74799.1 GGMS01003091 MBY72294.1 GFTR01004381 JAW12045.1 GECU01023166 GECU01011302 JAS84540.1 JAS96404.1 NEVH01021939 PNF19141.1 GBBI01001004 JAC17708.1 KQ434973 KZC12772.1 KT943559 ALQ33317.1 GECU01017682 JAS90024.1 ACPB03026421 GGMR01000597 MBY13216.1 KQ774368 OAD52321.1 LBMM01005052 KMQ91855.1 JRES01000262 KNC32891.1 ACPB03022475 GFXV01002129 MBW13934.1 KK852693 KDR18315.1 GL448949 EFN83556.1 JR052704 AEY61761.1 KQ977477 KYN02476.1 GFDF01002439 JAV11645.1 KQ982934 KYQ49138.1 LBMM01006207 KMQ90808.1 KQ414624 KOC67553.1 KOC67554.1 GL437663 EFN70139.1 CH954180 EDV47365.1 MG434621 ATY51927.1
KOB76812.1 RSAL01000008 RVE54009.1 AGBW02007651 OWR54902.1 KQ460878 KPJ11611.1 MG434602 ATY51908.1 KQ459582 KPI98834.1 MG434666 ATY51972.1 ODYU01000795 SOQ36115.1 NWSH01001518 PCG70999.1 BT128046 AEE63007.1 AXCN02000044 ADMH02001893 ETN60608.1 AXCM01013815 AAAB01008844 EAA06063.5 APCN01003147 GGFK01012571 MBW45892.1 GGFJ01008107 MBW57248.1 GGFJ01008108 MBW57249.1 CH477709 EAT37012.1 ATLV01019017 ATLV01019018 KE525259 KFB43518.1 DS235877 EEB19783.1 GAPW01002962 JAC10636.1 JXUM01028264 JXUM01030655 KQ560891 KQ560816 KXJ80465.1 KXJ80771.1 UFQS01000022 UFQT01000022 SSW97610.1 SSX17996.1 GGFJ01007550 MBW56691.1 GFDF01010299 JAV03785.1 GFDF01010667 JAV03417.1 PYGN01002057 PSN31652.1 GEBQ01025852 JAT14125.1 GEBQ01012094 JAT27883.1 AJWK01016944 AJWK01016945 AJWK01016946 AJWK01016947 AJWK01016948 AJWK01016949 CVRI01000057 CRL02023.1 GBBI01001854 JAC16858.1 GBGD01002381 JAC86508.1 GFTR01004631 JAW11795.1 GECL01003431 JAP02693.1 DS231832 EDS31789.1 GEDC01022556 GEDC01010710 GEDC01003689 GEDC01002746 JAS14742.1 JAS26588.1 JAS33609.1 JAS34552.1 EEB19784.1 GEBQ01018603 JAT21374.1 GEBQ01023362 JAT16615.1 GEZM01098136 JAV53982.1 GEDC01006573 JAS30725.1 GDKW01002174 JAI54421.1 GAHY01000155 JAA77355.1 KQ971312 EEZ98182.1 GFDL01005015 JAV30030.1 GEBQ01001890 JAT38087.1 GEDC01013602 JAS23696.1 GEBQ01003498 JAT36479.1 GEZM01051567 JAV74799.1 GGMS01003091 MBY72294.1 GFTR01004381 JAW12045.1 GECU01023166 GECU01011302 JAS84540.1 JAS96404.1 NEVH01021939 PNF19141.1 GBBI01001004 JAC17708.1 KQ434973 KZC12772.1 KT943559 ALQ33317.1 GECU01017682 JAS90024.1 ACPB03026421 GGMR01000597 MBY13216.1 KQ774368 OAD52321.1 LBMM01005052 KMQ91855.1 JRES01000262 KNC32891.1 ACPB03022475 GFXV01002129 MBW13934.1 KK852693 KDR18315.1 GL448949 EFN83556.1 JR052704 AEY61761.1 KQ977477 KYN02476.1 GFDF01002439 JAV11645.1 KQ982934 KYQ49138.1 LBMM01006207 KMQ90808.1 KQ414624 KOC67553.1 KOC67554.1 GL437663 EFN70139.1 CH954180 EDV47365.1 MG434621 ATY51927.1
Proteomes
UP000005204
UP000037510
UP000283053
UP000007151
UP000053240
UP000053268
+ More
UP000218220 UP000075886 UP000075920 UP000075880 UP000075900 UP000000673 UP000076408 UP000075883 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075885 UP000075884 UP000069272 UP000075903 UP000008820 UP000030765 UP000075881 UP000009046 UP000069940 UP000249989 UP000245037 UP000092461 UP000183832 UP000002320 UP000192223 UP000007266 UP000235965 UP000076502 UP000015103 UP000192221 UP000002358 UP000036403 UP000037069 UP000027135 UP000008237 UP000078542 UP000075809 UP000053825 UP000000311 UP000008711
UP000218220 UP000075886 UP000075920 UP000075880 UP000075900 UP000000673 UP000076408 UP000075883 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075885 UP000075884 UP000069272 UP000075903 UP000008820 UP000030765 UP000075881 UP000009046 UP000069940 UP000249989 UP000245037 UP000092461 UP000183832 UP000002320 UP000192223 UP000007266 UP000235965 UP000076502 UP000015103 UP000192221 UP000002358 UP000036403 UP000037069 UP000027135 UP000008237 UP000078542 UP000075809 UP000053825 UP000000311 UP000008711
PRIDE
Pfam
PF00650 CRAL_TRIO
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JTP5
A0A0S2Z3D7
A0A2W1C113
A0A0L7LMY9
A0A437BV40
A0A212FMC8
+ More
A0A194R6X1 A0A2H4RMP9 A0A194Q1G6 A0A2H4RMW5 A0A2H1V5I9 A0A2A4JGQ4 J3JXS5 A0A182QUC4 A0A182VY11 A0A182IJU1 A0A182R5Q8 W5JB24 A0A182YP37 A0A182LYI0 A0A182TXX4 A0A182WVC3 A0A182LAZ2 Q7QG15 A0A182I904 A0A2M4AYU9 A0A182PKF8 A0A182N4K3 A0A2M4BWZ8 A0A182F947 A0A2M4BW08 A0A182UZY6 Q16RH7 A0A084VZX4 A0A182K9E8 E0W2C7 A0A023ENV8 A0A182H7H8 A0A336K9A5 A0A2M4BUS0 A0A1L8DBN2 A0A1L8DAM2 A0A2P8XI41 A0A1B6KRS7 A0A1B6LW17 A0A1B0CL60 A0A1J1IP62 A0A023F6J1 A0A069DR11 A0A224XH32 A0A0V0G5X2 B0W3K3 A0A1B6DLR3 E0W2C8 A0A1B6LD20 A0A1B6KYX9 A0A1W4X5F9 A0A1Y1JXE4 A0A1B6DYJ3 A0A0P4VRB9 R4G5W8 D6W9C3 A0A1Q3FQZ8 A0A1B6MQ64 A0A1B6DDH3 A0A1B6MKT4 A0A1Y1LMD7 A0A2S2Q3J3 A0A224XHT0 A0A1B6JB60 A0A2J7PS36 A0A023F9V9 A0A154PLP4 A0A0S2Z365 A0A1B6ISW4 T1HQM6 A0A1W4VFQ6 K7ITY7 A0A2S2N8X6 A0A310SB26 A0A0J7NH54 A0A0L0CKW7 T1HWT6 A0A2H8TII4 A0A067R4X7 E2BL70 V9IMD6 A0A151IIH9 A0A1L8DZD6 A0A151WMV2 K7J0H5 A0A0J7KK55 A0A0L7R9L5 A0A0L7R9P5 E2A8R0 B3NXR4 A0A2H4RMS2
A0A194R6X1 A0A2H4RMP9 A0A194Q1G6 A0A2H4RMW5 A0A2H1V5I9 A0A2A4JGQ4 J3JXS5 A0A182QUC4 A0A182VY11 A0A182IJU1 A0A182R5Q8 W5JB24 A0A182YP37 A0A182LYI0 A0A182TXX4 A0A182WVC3 A0A182LAZ2 Q7QG15 A0A182I904 A0A2M4AYU9 A0A182PKF8 A0A182N4K3 A0A2M4BWZ8 A0A182F947 A0A2M4BW08 A0A182UZY6 Q16RH7 A0A084VZX4 A0A182K9E8 E0W2C7 A0A023ENV8 A0A182H7H8 A0A336K9A5 A0A2M4BUS0 A0A1L8DBN2 A0A1L8DAM2 A0A2P8XI41 A0A1B6KRS7 A0A1B6LW17 A0A1B0CL60 A0A1J1IP62 A0A023F6J1 A0A069DR11 A0A224XH32 A0A0V0G5X2 B0W3K3 A0A1B6DLR3 E0W2C8 A0A1B6LD20 A0A1B6KYX9 A0A1W4X5F9 A0A1Y1JXE4 A0A1B6DYJ3 A0A0P4VRB9 R4G5W8 D6W9C3 A0A1Q3FQZ8 A0A1B6MQ64 A0A1B6DDH3 A0A1B6MKT4 A0A1Y1LMD7 A0A2S2Q3J3 A0A224XHT0 A0A1B6JB60 A0A2J7PS36 A0A023F9V9 A0A154PLP4 A0A0S2Z365 A0A1B6ISW4 T1HQM6 A0A1W4VFQ6 K7ITY7 A0A2S2N8X6 A0A310SB26 A0A0J7NH54 A0A0L0CKW7 T1HWT6 A0A2H8TII4 A0A067R4X7 E2BL70 V9IMD6 A0A151IIH9 A0A1L8DZD6 A0A151WMV2 K7J0H5 A0A0J7KK55 A0A0L7R9L5 A0A0L7R9P5 E2A8R0 B3NXR4 A0A2H4RMS2
PDB
4CIZ
E-value=3.24173e-10,
Score=155
Ontologies
GO
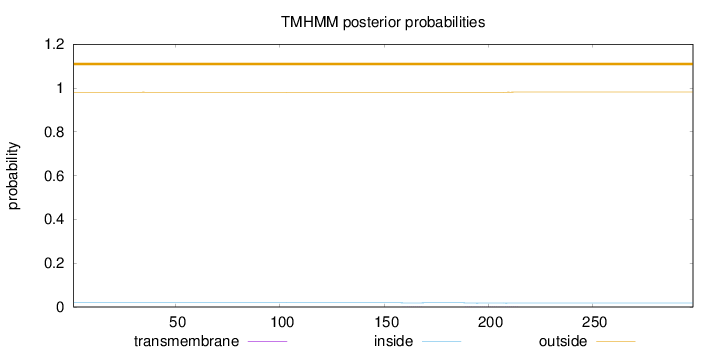
Topology
Length:
298
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02898
Exp number, first 60 AAs:
0.00074
Total prob of N-in:
0.01887
outside
1 - 298
Population Genetic Test Statistics
Pi
296.675396
Theta
157.359206
Tajima's D
2.614728
CLR
0.281698
CSRT
0.953902304884756
Interpretation
Uncertain
