Gene
KWMTBOMO09753
Pre Gene Modal
BGIBMGA012932
Annotation
PREDICTED:_leucine-zipper-like_transcriptional_regulator_1_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 1.222
Sequence
CDS
ATGCAGAACATTGCAAAGATGATAACAGAACAACTACGGAACCCGAATGAACTGGATATTAGCCTACGAATGGAGTTTGGACCTTTTGAGACTGTTCACAAATGGAAACGCATGTCTGAATGCTATGAATTTGTGGGTGCCAAGCGTAGTAAACACACAGCAATCGCTTATAAAGATGCAATATATGTGTTTGGAGGTGACAATGGAAAGTCAATGTTGAATGATCTGATCAGGTTTGATATAAGAGAGAAATCATGGACTAAAATGGGCTGTATGGGTATACCTCCAGCCCCAAGATATCATCATTCAGCTGTGGTACATCGTTCATCAATGTTTGTGTTTGGAGGCTATACTGGTGATATACTGGCAAACTCGAATCTAACAAATAAAAACGATCTCTTTGAGTACAAGTTCCAGAATGCACAATGGGTGCGGTGGAAGTTTACTGGACAGGAGCCAGTACCAAGATCAGCACACGGAGCTGCTGTGTATGATGACAAGCTGTGGATCTTTGCAGGTTATGATGGGAATGCCCGTCTTAACGACATGTGGACTATCGATTTAGTGGGAGAAACTCATCAATGGGAAAAAGTAGAACAGAAAGGTGAATGCCCTCCTACATGTTGCAACTTTCCTGTGGCTGTTGCTCGAGGGAAGATGTTTGTATTCAGCGGACAAAGTGGTGCTAAGATTACCAATGCCCTTTTCCAATTCGACTTTGAAAGTCGCATTTGGACCAGGGTGTGTACGGAGCATCTGCTGAGGTGTGCGGGGCCGGCCCCGGCGCGGCGCTACGGGCACGTAATGGCGGCGCACGGGCGACATCTGTACGTGTTTGGGGGGGCAGCCGATAACACGTTGCCCGCCGATTTGCACTGTTACGATCTCGATACGCAGATTTGGTCAGTTGTAAATCCGGCTCCCGATTCGCAAGTACCATCAGGGCGACTGTTCCACGCGGGCGCTGTCGTGGGCGACGCCTTGTACATCTTCGGTGGCACTGTTGATAACAACGTTAGGAGCGGAGAACTGTTTCGATTTCAGCTATCCGACTACCCTCGCTGCACCTTACATGAGGACTTCGGTCGCATTCTGAAATCGCAGCAGTTCTGCGACGTAACGTTACTGGTCGGAGACGAACAGACACCGGTACTAGCGCACCAAGCCATGTTGGCTGCCAGGTCTCAGTACCTCAGGACCAAAATAAAGGAAGCCCGTGAGGAACTATTGAGTAAGATAGAATCGGGCGAAGAGGCGGCCTCAAATCCATATTCGTACGACTCCCCGCCGCAGCTCACTGTAACACTCACCGAGGCCACACCAGAGGCGTTCAATATGGTACTCGACTACATATACACGGATAGGATTGATCCCACAGAGAAGGACGAGGACCCCGCGTCGCCGGCCACGATCCTGCTGGTGATGGAGGTGCTGCGGCTGGCGCTACGGCTGAGCATCCCGCGGCTGCGCGGCCTGTGCGCCCGCTTCCTGCGCGCCAACCTGTGCGCCACCAACGTGCTGCAGGCGCTGCACGCCGCGCACCACGCCGCCCTGCACTGCATCAAGGAGTATTGTCTCAGGTTCGTAGTGAAAGACTACAACTTCACGCCCATCGTGATGTCGCCTGAATTCGAACAGCTGGAGCAGAGCTTGATGGTGGAGGTGATCAGGAGGCGGCAGCAGCCGTCGTCCAAACAGGCAGCCCCGCACGACTCTGACGATGAGATCACGGGCACGACGCTGGAGCAGGACATGTGCGGGCTGGCGGGCGCGGGCGGCGCGGCGCTGAGCGACGTGCGGCTGCGCGCGGCGTCGCACGAGCGGCCCGCGCACCGCTCCATCCTGGCCGCGCGCGCCGCCTACTTCGAGGCCATGTTCCGCTCCTTCTCGCCGCGAGACAACATCGTCAATATACAGATATGCGACACAATTCCGTCGGAGGCAGCGTTCGACTCTCTCCTGCGCTACATCTACTACGGCGACACGAACATGCCCACAGAAGACTCTCTATATCTCTTCCAAGCTCCCATTTATTACGGATTCACAAACAATCGTCTCCAAGTGTTCTGCAAGCACAATCTACAGAGCAACGTGTCCCCTGAGAACGTGCTCGCCATACTGCAGGTCGCGGACAGGATGAAGGCGGCCGACATCAAAGAATACGCACTTAAAATGATCGTGCACCATTTTGGGGTTGTGGCGAGGCAAGAGGCAATAAAGAATCTAGGTCAACCGCTGCTGGTCGACATAATCTGGGCGCTCGCGGAGGAGCCGCAGTTCGAGGTGCCCCTCCCGGTCCAGCCGCACTCGCTGCCCTCCTCGTCCTCGGCAGATACGATCACGGACGACGCGGACTACGCGCGCAACAAGTGCAAGCCGCCGCACCGACTGCGCCATCCGGACTACAGCCTCGCCATCTGGCGCAGTAACAGCGATTAG
Protein
MQNIAKMITEQLRNPNELDISLRMEFGPFETVHKWKRMSECYEFVGAKRSKHTAIAYKDAIYVFGGDNGKSMLNDLIRFDIREKSWTKMGCMGIPPAPRYHHSAVVHRSSMFVFGGYTGDILANSNLTNKNDLFEYKFQNAQWVRWKFTGQEPVPRSAHGAAVYDDKLWIFAGYDGNARLNDMWTIDLVGETHQWEKVEQKGECPPTCCNFPVAVARGKMFVFSGQSGAKITNALFQFDFESRIWTRVCTEHLLRCAGPAPARRYGHVMAAHGRHLYVFGGAADNTLPADLHCYDLDTQIWSVVNPAPDSQVPSGRLFHAGAVVGDALYIFGGTVDNNVRSGELFRFQLSDYPRCTLHEDFGRILKSQQFCDVTLLVGDEQTPVLAHQAMLAARSQYLRTKIKEAREELLSKIESGEEAASNPYSYDSPPQLTVTLTEATPEAFNMVLDYIYTDRIDPTEKDEDPASPATILLVMEVLRLALRLSIPRLRGLCARFLRANLCATNVLQALHAAHHAALHCIKEYCLRFVVKDYNFTPIVMSPEFEQLEQSLMVEVIRRRQQPSSKQAAPHDSDDEITGTTLEQDMCGLAGAGGAALSDVRLRAASHERPAHRSILAARAAYFEAMFRSFSPRDNIVNIQICDTIPSEAAFDSLLRYIYYGDTNMPTEDSLYLFQAPIYYGFTNNRLQVFCKHNLQSNVSPENVLAILQVADRMKAADIKEYALKMIVHHFGVVARQEAIKNLGQPLLVDIIWALAEEPQFEVPLPVQPHSLPSSSSADTITDDADYARNKCKPPHRLRHPDYSLAIWRSNSD
Summary
Uniprot
H9JTS0
A0A212FMF1
A0A3S2M9G9
A0A194R6X5
A0A2A4K2Y6
A0A2P8YAL2
+ More
A0A088ASA1 A0A1W4XCW3 K7IML4 A0A0L7RH49 A0A0M9A479 E2BZW3 A0A2J7R857 A0A067R7N5 A0A154P834 A0A1Y1K1T1 T1JJ12 A0A232EV99 A0A3R7M9Y3 A0A0J7L4I8 A0A224YSJ5 A0A131YY60 L7M727 E2AF29 A0A1Q3G2Z3 A0A0P6EQB8 E9GTI5 A0A1L8DFM9 A0A0P6IFG7 A0A131XAC3 A0A0P5S2L8 A0A162SH89 A0A0P5NA27 A0A026WAV4 A0A0P4VUF9 A0A293M809 A0A1S4EX93 A0A182FR40 Q17MW2 W5JEM8 A0A084VKC5 A0A1S3JT25 A0A0N8E222 A0A182IR83 A0A182R5L8 A0A182X8V4 A0A182PRG9 A0A0N8BGY9 A0A182VY56 A0A182M466 A0A2R5LLC3 A0A1S3JS18 A0A182UY76 A0A182TJE7 A0A182L6W8 Q7QAD5 A0A182ID36 A0A182HCR8 A0A182ND46 A0A182QIK2 A0A0P5E7N5 A0A182JPE0 A0A2A3EM45 A0A182XWQ3 A0A0N8BJX6 V5GXH1 A0A2M4BFC4 E9I9G1 D2A1P2 A0A0P5MXF5 V4A003 A0A0P4Y6D2 A0A195FU41 A0A158NA56 A0A1B0CY87 A0A195BBI7 A0A401SXF0 W5M8F1 B4ND63 A0A1W4Z5M6 R7TR27 A0A151N8N7 A0A3B0KR67 A0A034VIQ8 B4MD42 Q6NRT1 Q29FN7 A0A1W4YWR4 A0A3B4ECD8 W8AVL0 A0A1L8HQX2 A0A2R8QN10 B2GUN2 A9JRR0 B3MYC8 F1R533 A0A0B8RQB0 A0A098LZ46 W5K0V8
A0A088ASA1 A0A1W4XCW3 K7IML4 A0A0L7RH49 A0A0M9A479 E2BZW3 A0A2J7R857 A0A067R7N5 A0A154P834 A0A1Y1K1T1 T1JJ12 A0A232EV99 A0A3R7M9Y3 A0A0J7L4I8 A0A224YSJ5 A0A131YY60 L7M727 E2AF29 A0A1Q3G2Z3 A0A0P6EQB8 E9GTI5 A0A1L8DFM9 A0A0P6IFG7 A0A131XAC3 A0A0P5S2L8 A0A162SH89 A0A0P5NA27 A0A026WAV4 A0A0P4VUF9 A0A293M809 A0A1S4EX93 A0A182FR40 Q17MW2 W5JEM8 A0A084VKC5 A0A1S3JT25 A0A0N8E222 A0A182IR83 A0A182R5L8 A0A182X8V4 A0A182PRG9 A0A0N8BGY9 A0A182VY56 A0A182M466 A0A2R5LLC3 A0A1S3JS18 A0A182UY76 A0A182TJE7 A0A182L6W8 Q7QAD5 A0A182ID36 A0A182HCR8 A0A182ND46 A0A182QIK2 A0A0P5E7N5 A0A182JPE0 A0A2A3EM45 A0A182XWQ3 A0A0N8BJX6 V5GXH1 A0A2M4BFC4 E9I9G1 D2A1P2 A0A0P5MXF5 V4A003 A0A0P4Y6D2 A0A195FU41 A0A158NA56 A0A1B0CY87 A0A195BBI7 A0A401SXF0 W5M8F1 B4ND63 A0A1W4Z5M6 R7TR27 A0A151N8N7 A0A3B0KR67 A0A034VIQ8 B4MD42 Q6NRT1 Q29FN7 A0A1W4YWR4 A0A3B4ECD8 W8AVL0 A0A1L8HQX2 A0A2R8QN10 B2GUN2 A9JRR0 B3MYC8 F1R533 A0A0B8RQB0 A0A098LZ46 W5K0V8
Pubmed
19121390
22118469
26354079
29403074
20075255
20798317
+ More
24845553 28004739 28648823 28797301 26830274 25576852 21292972 28049606 24508170 30249741 17510324 20920257 23761445 24438588 20966253 12364791 14747013 17210077 26483478 25244985 25765539 21282665 18362917 19820115 23254933 21347285 30297745 17994087 22293439 25348373 15632085 24495485 27762356 23594743 25476704 25329095
24845553 28004739 28648823 28797301 26830274 25576852 21292972 28049606 24508170 30249741 17510324 20920257 23761445 24438588 20966253 12364791 14747013 17210077 26483478 25244985 25765539 21282665 18362917 19820115 23254933 21347285 30297745 17994087 22293439 25348373 15632085 24495485 27762356 23594743 25476704 25329095
EMBL
BABH01003432
BABH01003433
AGBW02007651
OWR54907.1
RSAL01000008
RVE54016.1
+ More
KQ460878 KPJ11616.1 NWSH01000188 PCG78615.1 PYGN01000753 PSN41296.1 KQ414590 KOC70302.1 KQ435753 KOX76136.1 GL451708 EFN78740.1 NEVH01006723 PNF37011.1 KK852685 KDR18486.1 KQ434839 KZC08032.1 GEZM01099430 JAV53316.1 JH431859 NNAY01002028 OXU22270.1 QCYY01001697 ROT76051.1 LBMM01000826 KMQ97409.1 GFPF01009431 MAA20577.1 GEDV01004630 JAP83927.1 GACK01005179 JAA59855.1 GL438984 EFN67984.1 GFDL01000873 JAV34172.1 GDIQ01185341 GDIQ01175350 GDIQ01153410 GDIQ01113630 GDIP01053645 GDIQ01080395 GDIQ01078486 GDIQ01072890 GDIQ01059894 JAM50070.1 JAN21847.1 GL732564 EFX77089.1 GFDF01008843 JAV05241.1 GDIQ01005331 JAN89406.1 GEFH01004568 JAP64013.1 GDIQ01093184 JAL58542.1 LRGB01000024 KZS21295.1 GDIQ01156058 GDIQ01156055 JAK95667.1 KK107320 QOIP01000009 EZA52806.1 RLU18506.1 GDRN01095806 GDRN01095805 JAI59454.1 GFWV01010541 MAA35270.1 CH477203 EAT48040.1 ADMH02001710 ETN61345.1 ATLV01014178 KE524947 KFB38419.1 GDIQ01069492 JAN25245.1 GDIQ01178747 GDIQ01039948 JAK72978.1 AXCM01005876 GGLE01006176 MBY10302.1 AAAB01008898 EAA09185.5 APCN01004985 JXUM01034490 KQ561024 KXJ79986.1 AXCN02001581 GDIP01150477 JAJ72925.1 KZ288218 PBC32352.1 GDIQ01170451 JAK81274.1 GANP01009273 JAB75195.1 GGFJ01002603 MBW51744.1 GL761800 EFZ22805.1 KQ971338 EFA02704.1 GDIQ01150418 JAL01308.1 KB202518 ESO89967.1 GDIP01232926 JAI90475.1 KQ981264 KYN43978.1 ADTU01001427 AJWK01035581 KQ976528 KYM81898.1 BEZZ01000663 GCC35087.1 AHAT01005101 CH964239 EDW82772.2 AMQN01012506 KB309561 ELT93946.1 AKHW03003787 KYO33193.1 OUUW01000018 SPP89129.1 GAKP01016573 JAC42379.1 CH940660 EDW58114.2 BC070638 AAH70638.1 CH379065 EAL31542.3 GAMC01017812 GAMC01017810 JAB88743.1 CM004467 OCT98485.1 CR855305 BC166344 AAI66344.1 CT737234 BC155757 AAI55758.1 CH902632 EDV32622.2 GBSH01001472 JAG67554.1 GBSI01001260 JAC95236.1
KQ460878 KPJ11616.1 NWSH01000188 PCG78615.1 PYGN01000753 PSN41296.1 KQ414590 KOC70302.1 KQ435753 KOX76136.1 GL451708 EFN78740.1 NEVH01006723 PNF37011.1 KK852685 KDR18486.1 KQ434839 KZC08032.1 GEZM01099430 JAV53316.1 JH431859 NNAY01002028 OXU22270.1 QCYY01001697 ROT76051.1 LBMM01000826 KMQ97409.1 GFPF01009431 MAA20577.1 GEDV01004630 JAP83927.1 GACK01005179 JAA59855.1 GL438984 EFN67984.1 GFDL01000873 JAV34172.1 GDIQ01185341 GDIQ01175350 GDIQ01153410 GDIQ01113630 GDIP01053645 GDIQ01080395 GDIQ01078486 GDIQ01072890 GDIQ01059894 JAM50070.1 JAN21847.1 GL732564 EFX77089.1 GFDF01008843 JAV05241.1 GDIQ01005331 JAN89406.1 GEFH01004568 JAP64013.1 GDIQ01093184 JAL58542.1 LRGB01000024 KZS21295.1 GDIQ01156058 GDIQ01156055 JAK95667.1 KK107320 QOIP01000009 EZA52806.1 RLU18506.1 GDRN01095806 GDRN01095805 JAI59454.1 GFWV01010541 MAA35270.1 CH477203 EAT48040.1 ADMH02001710 ETN61345.1 ATLV01014178 KE524947 KFB38419.1 GDIQ01069492 JAN25245.1 GDIQ01178747 GDIQ01039948 JAK72978.1 AXCM01005876 GGLE01006176 MBY10302.1 AAAB01008898 EAA09185.5 APCN01004985 JXUM01034490 KQ561024 KXJ79986.1 AXCN02001581 GDIP01150477 JAJ72925.1 KZ288218 PBC32352.1 GDIQ01170451 JAK81274.1 GANP01009273 JAB75195.1 GGFJ01002603 MBW51744.1 GL761800 EFZ22805.1 KQ971338 EFA02704.1 GDIQ01150418 JAL01308.1 KB202518 ESO89967.1 GDIP01232926 JAI90475.1 KQ981264 KYN43978.1 ADTU01001427 AJWK01035581 KQ976528 KYM81898.1 BEZZ01000663 GCC35087.1 AHAT01005101 CH964239 EDW82772.2 AMQN01012506 KB309561 ELT93946.1 AKHW03003787 KYO33193.1 OUUW01000018 SPP89129.1 GAKP01016573 JAC42379.1 CH940660 EDW58114.2 BC070638 AAH70638.1 CH379065 EAL31542.3 GAMC01017812 GAMC01017810 JAB88743.1 CM004467 OCT98485.1 CR855305 BC166344 AAI66344.1 CT737234 BC155757 AAI55758.1 CH902632 EDV32622.2 GBSH01001472 JAG67554.1 GBSI01001260 JAC95236.1
Proteomes
UP000005204
UP000007151
UP000283053
UP000053240
UP000218220
UP000245037
+ More
UP000005203 UP000192223 UP000002358 UP000053825 UP000053105 UP000008237 UP000235965 UP000027135 UP000076502 UP000215335 UP000283509 UP000036403 UP000000311 UP000000305 UP000076858 UP000053097 UP000279307 UP000069272 UP000008820 UP000000673 UP000030765 UP000085678 UP000075880 UP000075900 UP000076407 UP000075885 UP000075920 UP000075883 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840 UP000069940 UP000249989 UP000075884 UP000075886 UP000075881 UP000242457 UP000076408 UP000007266 UP000030746 UP000078541 UP000005205 UP000092461 UP000078540 UP000287033 UP000018468 UP000007798 UP000192224 UP000014760 UP000050525 UP000268350 UP000008792 UP000001819 UP000261440 UP000186698 UP000000437 UP000007801 UP000018467
UP000005203 UP000192223 UP000002358 UP000053825 UP000053105 UP000008237 UP000235965 UP000027135 UP000076502 UP000215335 UP000283509 UP000036403 UP000000311 UP000000305 UP000076858 UP000053097 UP000279307 UP000069272 UP000008820 UP000000673 UP000030765 UP000085678 UP000075880 UP000075900 UP000076407 UP000075885 UP000075920 UP000075883 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840 UP000069940 UP000249989 UP000075884 UP000075886 UP000075881 UP000242457 UP000076408 UP000007266 UP000030746 UP000078541 UP000005205 UP000092461 UP000078540 UP000287033 UP000018468 UP000007798 UP000192224 UP000014760 UP000050525 UP000268350 UP000008792 UP000001819 UP000261440 UP000186698 UP000000437 UP000007801 UP000018467
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JTS0
A0A212FMF1
A0A3S2M9G9
A0A194R6X5
A0A2A4K2Y6
A0A2P8YAL2
+ More
A0A088ASA1 A0A1W4XCW3 K7IML4 A0A0L7RH49 A0A0M9A479 E2BZW3 A0A2J7R857 A0A067R7N5 A0A154P834 A0A1Y1K1T1 T1JJ12 A0A232EV99 A0A3R7M9Y3 A0A0J7L4I8 A0A224YSJ5 A0A131YY60 L7M727 E2AF29 A0A1Q3G2Z3 A0A0P6EQB8 E9GTI5 A0A1L8DFM9 A0A0P6IFG7 A0A131XAC3 A0A0P5S2L8 A0A162SH89 A0A0P5NA27 A0A026WAV4 A0A0P4VUF9 A0A293M809 A0A1S4EX93 A0A182FR40 Q17MW2 W5JEM8 A0A084VKC5 A0A1S3JT25 A0A0N8E222 A0A182IR83 A0A182R5L8 A0A182X8V4 A0A182PRG9 A0A0N8BGY9 A0A182VY56 A0A182M466 A0A2R5LLC3 A0A1S3JS18 A0A182UY76 A0A182TJE7 A0A182L6W8 Q7QAD5 A0A182ID36 A0A182HCR8 A0A182ND46 A0A182QIK2 A0A0P5E7N5 A0A182JPE0 A0A2A3EM45 A0A182XWQ3 A0A0N8BJX6 V5GXH1 A0A2M4BFC4 E9I9G1 D2A1P2 A0A0P5MXF5 V4A003 A0A0P4Y6D2 A0A195FU41 A0A158NA56 A0A1B0CY87 A0A195BBI7 A0A401SXF0 W5M8F1 B4ND63 A0A1W4Z5M6 R7TR27 A0A151N8N7 A0A3B0KR67 A0A034VIQ8 B4MD42 Q6NRT1 Q29FN7 A0A1W4YWR4 A0A3B4ECD8 W8AVL0 A0A1L8HQX2 A0A2R8QN10 B2GUN2 A9JRR0 B3MYC8 F1R533 A0A0B8RQB0 A0A098LZ46 W5K0V8
A0A088ASA1 A0A1W4XCW3 K7IML4 A0A0L7RH49 A0A0M9A479 E2BZW3 A0A2J7R857 A0A067R7N5 A0A154P834 A0A1Y1K1T1 T1JJ12 A0A232EV99 A0A3R7M9Y3 A0A0J7L4I8 A0A224YSJ5 A0A131YY60 L7M727 E2AF29 A0A1Q3G2Z3 A0A0P6EQB8 E9GTI5 A0A1L8DFM9 A0A0P6IFG7 A0A131XAC3 A0A0P5S2L8 A0A162SH89 A0A0P5NA27 A0A026WAV4 A0A0P4VUF9 A0A293M809 A0A1S4EX93 A0A182FR40 Q17MW2 W5JEM8 A0A084VKC5 A0A1S3JT25 A0A0N8E222 A0A182IR83 A0A182R5L8 A0A182X8V4 A0A182PRG9 A0A0N8BGY9 A0A182VY56 A0A182M466 A0A2R5LLC3 A0A1S3JS18 A0A182UY76 A0A182TJE7 A0A182L6W8 Q7QAD5 A0A182ID36 A0A182HCR8 A0A182ND46 A0A182QIK2 A0A0P5E7N5 A0A182JPE0 A0A2A3EM45 A0A182XWQ3 A0A0N8BJX6 V5GXH1 A0A2M4BFC4 E9I9G1 D2A1P2 A0A0P5MXF5 V4A003 A0A0P4Y6D2 A0A195FU41 A0A158NA56 A0A1B0CY87 A0A195BBI7 A0A401SXF0 W5M8F1 B4ND63 A0A1W4Z5M6 R7TR27 A0A151N8N7 A0A3B0KR67 A0A034VIQ8 B4MD42 Q6NRT1 Q29FN7 A0A1W4YWR4 A0A3B4ECD8 W8AVL0 A0A1L8HQX2 A0A2R8QN10 B2GUN2 A9JRR0 B3MYC8 F1R533 A0A0B8RQB0 A0A098LZ46 W5K0V8
PDB
6DO5
E-value=2.47191e-16,
Score=212
Ontologies
GO
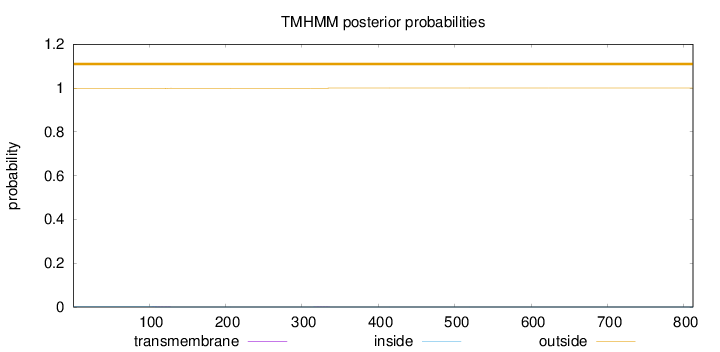
Topology
Length:
812
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0319099999999999
Exp number, first 60 AAs:
0.00017
Total prob of N-in:
0.00164
outside
1 - 812
Population Genetic Test Statistics
Pi
273.796938
Theta
174.48514
Tajima's D
1.756503
CLR
0.324662
CSRT
0.842557872106395
Interpretation
Uncertain
