Pre Gene Modal
BGIBMGA012934
Annotation
actin_related_protein_2/3_complex_subunit_4_[Bombyx_mori]
Full name
Actin-related protein 2/3 complex subunit 4
Location in the cell
Cytoplasmic Reliability : 1.29 Mitochondrial Reliability : 1.212
Sequence
CDS
ATGGCTGCAACTTTAAAACCGTATCTAACAGCGGTGCGTCACACGCTCACTGCCGCTATGTGCTTAGAACACTTTTCATCGCAAGTTGTCGAGAAATACACCAAGCCGGAGGTCGAAGTTAGAACAAGCAAAGAACTTTTACTCAATCCTGTTGTTATTTCAAGAAATTCTAATGAAAAAGTATTAATAGAGTCTTCAATAAATTCAATTAGGGTGAGTATTATGATCAAGCAGGCGGATGAGATAGAGAAAATTCTTTGTAAAAAATTTATGCGGTTCATGATGATGAGAGCTGAAAACTTCATTGTATTAAGGAGGAAACCAGTTGATGGGTACCATATCAGCTTTTTGATTACTAACTTCCATACCGAACAGATGTATAAGCATAAATTGGTCGATTTTGTAATATACTTTATGGAAGAAATTGATAAAGAAATCAGTGAAATGAAATTGGCTGTCAATGCAAGAGCCAGAATTTGTTCGGAAGAATTCTTGAAGAGATTTTAA
Protein
MAATLKPYLTAVRHTLTAAMCLEHFSSQVVEKYTKPEVEVRTSKELLLNPVVISRNSNEKVLIESSINSIRVSIMIKQADEIEKILCKKFMRFMMMRAENFIVLRRKPVDGYHISFLITNFHTEQMYKHKLVDFVIYFMEEIDKEISEMKLAVNARARICSEEFLKRF
Summary
Description
Functions as actin-binding component of the Arp2/3 complex which is involved in regulation of actin polymerization and together with an activating nucleation-promoting factor (NPF) mediates the formation of branched actin networks. Seems to contact the mother actin filament.
Similarity
Belongs to the ARPC4 family.
Uniprot
Q1HPT2
A0A2W1C0K9
A0A2A4K3J7
A0A0L7L8M3
A0A2H1V4T4
X4Z1B2
+ More
A0A3S2LUH6 A0A194R276 A0A194Q5Z2 A0A2P8XRL7 A0A0T6B152 A0A226EWT5 A0A182SJZ9 A0A182YBE0 A0A182PTS0 A0A182M1X8 A0A182Q4M7 A0A182NR76 A0A182VVT4 A0A182TVD0 A0A182X9K7 A0A182L8T5 Q7PUB7 A0A182RNJ7 A0A182HKJ0 A0A1D2NGH8 A0A182V4U4 A0A023EHV7 A0A1Y1MQE2 T1E8K8 A0A2M3Z8U7 A0A2M4APG7 W5J3R1 A0A084VXV3 A0A182IXM0 A0A182FRK7 D2A6A8 A0A1L8E5Q5 A0A1B0GI81 A0A1Q3FG03 B0X5P8 Q16WZ0 U5ELN7 A0A2L2Y2K6 T1DSL0 A0A0P5KDK0 A0A224YIR3 A0A0K8RH12 A0A212FMJ7 A0A1E1WWE6 A0A087U3N5 A0A1J1ITS5 A0A131Z4K4 L7LY75 A0A131XIW3 A0A023FHE6 S4PL32 A0A1B6D5I3 A0A1W4X8B9 A0A1E1XUH2 A0A023FEX5 A0A2A4K3Z8 G3MNL8 A0A1S3D024 A0A2R5LNJ4 A0A1B6J3U4 E0VA94 A0A0K8TLR8 A0A1B6LN36 A0A182K4E5 A0A023NL51 A0A2R7WRQ9 R7UGT8 A0A0L8FI18 A0A293M415 A0A3M6T475 A0A1B6HV00 A0A1Z5KUB3 A0A0K8SJF4 A7RRG4 N6UG83 A0A147BA47 T1F6L5 A0A3P8TMD2 A0A3B1JWY7 A0A3B4CCU3 A0A1S3JQ95 A0A1A8M9H1 A0A0E9X7D4 A0A1A8ATX8 R4WDL1 A0A1W4Z8H0 F7FPY5 W8C601 A0A0K8UZ82 A0A034VPQ5 A0A3B4XFD9 A0A3Q2P008 A0A3Q2DSM1 A0A3B3YZY1
A0A3S2LUH6 A0A194R276 A0A194Q5Z2 A0A2P8XRL7 A0A0T6B152 A0A226EWT5 A0A182SJZ9 A0A182YBE0 A0A182PTS0 A0A182M1X8 A0A182Q4M7 A0A182NR76 A0A182VVT4 A0A182TVD0 A0A182X9K7 A0A182L8T5 Q7PUB7 A0A182RNJ7 A0A182HKJ0 A0A1D2NGH8 A0A182V4U4 A0A023EHV7 A0A1Y1MQE2 T1E8K8 A0A2M3Z8U7 A0A2M4APG7 W5J3R1 A0A084VXV3 A0A182IXM0 A0A182FRK7 D2A6A8 A0A1L8E5Q5 A0A1B0GI81 A0A1Q3FG03 B0X5P8 Q16WZ0 U5ELN7 A0A2L2Y2K6 T1DSL0 A0A0P5KDK0 A0A224YIR3 A0A0K8RH12 A0A212FMJ7 A0A1E1WWE6 A0A087U3N5 A0A1J1ITS5 A0A131Z4K4 L7LY75 A0A131XIW3 A0A023FHE6 S4PL32 A0A1B6D5I3 A0A1W4X8B9 A0A1E1XUH2 A0A023FEX5 A0A2A4K3Z8 G3MNL8 A0A1S3D024 A0A2R5LNJ4 A0A1B6J3U4 E0VA94 A0A0K8TLR8 A0A1B6LN36 A0A182K4E5 A0A023NL51 A0A2R7WRQ9 R7UGT8 A0A0L8FI18 A0A293M415 A0A3M6T475 A0A1B6HV00 A0A1Z5KUB3 A0A0K8SJF4 A7RRG4 N6UG83 A0A147BA47 T1F6L5 A0A3P8TMD2 A0A3B1JWY7 A0A3B4CCU3 A0A1S3JQ95 A0A1A8M9H1 A0A0E9X7D4 A0A1A8ATX8 R4WDL1 A0A1W4Z8H0 F7FPY5 W8C601 A0A0K8UZ82 A0A034VPQ5 A0A3B4XFD9 A0A3Q2P008 A0A3Q2DSM1 A0A3B3YZY1
Pubmed
28756777
26227816
26354079
29403074
25244985
20966253
+ More
12364791 14747013 17210077 27289101 24945155 26483478 28004739 20920257 23761445 24438588 18362917 19820115 17510324 26561354 28797301 22118469 28503490 26830274 25576852 28049606 23622113 29209593 22216098 20566863 26369729 24733187 23254933 30382153 28528879 26823975 17615350 23537049 25329095 25613341 23691247 18464734 24495485 25348373
12364791 14747013 17210077 27289101 24945155 26483478 28004739 20920257 23761445 24438588 18362917 19820115 17510324 26561354 28797301 22118469 28503490 26830274 25576852 28049606 23622113 29209593 22216098 20566863 26369729 24733187 23254933 30382153 28528879 26823975 17615350 23537049 25329095 25613341 23691247 18464734 24495485 25348373
EMBL
DQ443320
ABF51409.1
KZ149898
PZC78650.1
NWSH01000188
PCG78629.1
+ More
JTDY01002320 KOB71664.1 ODYU01000678 SOQ35860.1 KJ187401 AHV90276.1 RSAL01000008 RVE54019.1 KQ460878 KPJ11619.1 KQ459582 KPI98825.1 PYGN01001468 PSN34646.1 LJIG01016367 KRT80814.1 LNIX01000001 OXA61650.1 AXCM01001747 AXCN02001469 AAAB01008987 EAA01786.3 APCN01000833 LJIJ01000056 ODN04066.1 JXUM01000125 JXUM01055969 GAPW01005137 KQ561891 KQ560101 JAC08461.1 KXJ77244.1 KXJ84563.1 GEZM01027049 JAV86850.1 GAMD01002678 JAA98912.1 GGFM01004206 MBW24957.1 GGFK01009326 MBW42647.1 ADMH02002105 GGFL01004229 ETN59007.1 MBW68407.1 ATLV01018208 KE525224 KFB42797.1 AXCP01002658 KQ971347 EFA05486.1 GFDF01000125 JAV13959.1 AJWK01013570 GFDL01008566 JAV26479.1 DS232391 EDS40985.1 CH477549 EAT39131.1 GANO01001329 JAB58542.1 IAAA01004181 LAA02402.1 GAMD01000150 JAB01441.1 GDIQ01192109 GDIP01068718 LRGB01001005 JAK59616.1 JAM34997.1 KZS14177.1 GFPF01005709 MAA16855.1 GADI01003378 JAA70430.1 AGBW02007651 OWR54909.1 GFAC01007835 JAT91353.1 KK118013 KFM71974.1 CVRI01000057 CRL01921.1 GEDV01002649 JAP85908.1 GACK01008294 JAA56740.1 GEFH01002279 JAP66302.1 GBBK01004027 JAC20455.1 GAIX01000781 JAA91779.1 GEDC01016356 JAS20942.1 GFAA01000658 JAU02777.1 GBBK01004026 JAC20456.1 PCG78628.1 JO843469 AEO35086.1 GGLE01006899 MBY11025.1 GECU01027313 GECU01013860 JAS80393.1 JAS93846.1 DS235005 EEB10300.1 GDAI01002728 JAI14875.1 GEBQ01014988 JAT24989.1 KF780489 AHX02953.1 KK855372 PTY22246.1 AMQN01008807 KB304010 ELU02472.1 KQ431294 KOF63267.1 GFWV01010307 MAA35036.1 RCHS01004365 RMX35032.1 GECU01029214 GECU01011366 JAS78492.1 JAS96340.1 GFJQ02008278 JAV98691.1 GBRD01012391 GDHC01021963 JAG53433.1 JAP96665.1 DS469531 EDO46006.1 APGK01036710 KB740941 KB632225 ENN77657.1 ERL90283.1 GEIB01000745 JAR87245.1 AMQM01004492 KB096590 ESO03545.1 HAEF01012052 SBR53211.1 GBXM01009915 JAH98662.1 HADY01019210 SBP57695.1 AK417624 BAN20839.1 GAMC01004594 JAC01962.1 GDHF01020332 GDHF01007226 JAI31982.1 JAI45088.1 GAKP01015167 GAKP01015165 GAKP01015163 JAC43785.1
JTDY01002320 KOB71664.1 ODYU01000678 SOQ35860.1 KJ187401 AHV90276.1 RSAL01000008 RVE54019.1 KQ460878 KPJ11619.1 KQ459582 KPI98825.1 PYGN01001468 PSN34646.1 LJIG01016367 KRT80814.1 LNIX01000001 OXA61650.1 AXCM01001747 AXCN02001469 AAAB01008987 EAA01786.3 APCN01000833 LJIJ01000056 ODN04066.1 JXUM01000125 JXUM01055969 GAPW01005137 KQ561891 KQ560101 JAC08461.1 KXJ77244.1 KXJ84563.1 GEZM01027049 JAV86850.1 GAMD01002678 JAA98912.1 GGFM01004206 MBW24957.1 GGFK01009326 MBW42647.1 ADMH02002105 GGFL01004229 ETN59007.1 MBW68407.1 ATLV01018208 KE525224 KFB42797.1 AXCP01002658 KQ971347 EFA05486.1 GFDF01000125 JAV13959.1 AJWK01013570 GFDL01008566 JAV26479.1 DS232391 EDS40985.1 CH477549 EAT39131.1 GANO01001329 JAB58542.1 IAAA01004181 LAA02402.1 GAMD01000150 JAB01441.1 GDIQ01192109 GDIP01068718 LRGB01001005 JAK59616.1 JAM34997.1 KZS14177.1 GFPF01005709 MAA16855.1 GADI01003378 JAA70430.1 AGBW02007651 OWR54909.1 GFAC01007835 JAT91353.1 KK118013 KFM71974.1 CVRI01000057 CRL01921.1 GEDV01002649 JAP85908.1 GACK01008294 JAA56740.1 GEFH01002279 JAP66302.1 GBBK01004027 JAC20455.1 GAIX01000781 JAA91779.1 GEDC01016356 JAS20942.1 GFAA01000658 JAU02777.1 GBBK01004026 JAC20456.1 PCG78628.1 JO843469 AEO35086.1 GGLE01006899 MBY11025.1 GECU01027313 GECU01013860 JAS80393.1 JAS93846.1 DS235005 EEB10300.1 GDAI01002728 JAI14875.1 GEBQ01014988 JAT24989.1 KF780489 AHX02953.1 KK855372 PTY22246.1 AMQN01008807 KB304010 ELU02472.1 KQ431294 KOF63267.1 GFWV01010307 MAA35036.1 RCHS01004365 RMX35032.1 GECU01029214 GECU01011366 JAS78492.1 JAS96340.1 GFJQ02008278 JAV98691.1 GBRD01012391 GDHC01021963 JAG53433.1 JAP96665.1 DS469531 EDO46006.1 APGK01036710 KB740941 KB632225 ENN77657.1 ERL90283.1 GEIB01000745 JAR87245.1 AMQM01004492 KB096590 ESO03545.1 HAEF01012052 SBR53211.1 GBXM01009915 JAH98662.1 HADY01019210 SBP57695.1 AK417624 BAN20839.1 GAMC01004594 JAC01962.1 GDHF01020332 GDHF01007226 JAI31982.1 JAI45088.1 GAKP01015167 GAKP01015165 GAKP01015163 JAC43785.1
Proteomes
UP000218220
UP000037510
UP000283053
UP000053240
UP000053268
UP000245037
+ More
UP000198287 UP000075901 UP000076408 UP000075885 UP000075883 UP000075886 UP000075884 UP000075920 UP000075902 UP000076407 UP000075882 UP000007062 UP000075900 UP000075840 UP000094527 UP000075903 UP000069940 UP000249989 UP000000673 UP000030765 UP000075880 UP000069272 UP000007266 UP000092461 UP000002320 UP000008820 UP000076858 UP000007151 UP000054359 UP000183832 UP000192223 UP000079169 UP000009046 UP000075881 UP000014760 UP000053454 UP000275408 UP000001593 UP000019118 UP000030742 UP000015101 UP000265080 UP000018467 UP000261440 UP000085678 UP000192224 UP000002279 UP000261360 UP000265000 UP000265020 UP000261480
UP000198287 UP000075901 UP000076408 UP000075885 UP000075883 UP000075886 UP000075884 UP000075920 UP000075902 UP000076407 UP000075882 UP000007062 UP000075900 UP000075840 UP000094527 UP000075903 UP000069940 UP000249989 UP000000673 UP000030765 UP000075880 UP000069272 UP000007266 UP000092461 UP000002320 UP000008820 UP000076858 UP000007151 UP000054359 UP000183832 UP000192223 UP000079169 UP000009046 UP000075881 UP000014760 UP000053454 UP000275408 UP000001593 UP000019118 UP000030742 UP000015101 UP000265080 UP000018467 UP000261440 UP000085678 UP000192224 UP000002279 UP000261360 UP000265000 UP000265020 UP000261480
Pfam
PF05856 ARPC4
SUPFAM
SSF69645
SSF69645
Gene 3D
ProteinModelPortal
Q1HPT2
A0A2W1C0K9
A0A2A4K3J7
A0A0L7L8M3
A0A2H1V4T4
X4Z1B2
+ More
A0A3S2LUH6 A0A194R276 A0A194Q5Z2 A0A2P8XRL7 A0A0T6B152 A0A226EWT5 A0A182SJZ9 A0A182YBE0 A0A182PTS0 A0A182M1X8 A0A182Q4M7 A0A182NR76 A0A182VVT4 A0A182TVD0 A0A182X9K7 A0A182L8T5 Q7PUB7 A0A182RNJ7 A0A182HKJ0 A0A1D2NGH8 A0A182V4U4 A0A023EHV7 A0A1Y1MQE2 T1E8K8 A0A2M3Z8U7 A0A2M4APG7 W5J3R1 A0A084VXV3 A0A182IXM0 A0A182FRK7 D2A6A8 A0A1L8E5Q5 A0A1B0GI81 A0A1Q3FG03 B0X5P8 Q16WZ0 U5ELN7 A0A2L2Y2K6 T1DSL0 A0A0P5KDK0 A0A224YIR3 A0A0K8RH12 A0A212FMJ7 A0A1E1WWE6 A0A087U3N5 A0A1J1ITS5 A0A131Z4K4 L7LY75 A0A131XIW3 A0A023FHE6 S4PL32 A0A1B6D5I3 A0A1W4X8B9 A0A1E1XUH2 A0A023FEX5 A0A2A4K3Z8 G3MNL8 A0A1S3D024 A0A2R5LNJ4 A0A1B6J3U4 E0VA94 A0A0K8TLR8 A0A1B6LN36 A0A182K4E5 A0A023NL51 A0A2R7WRQ9 R7UGT8 A0A0L8FI18 A0A293M415 A0A3M6T475 A0A1B6HV00 A0A1Z5KUB3 A0A0K8SJF4 A7RRG4 N6UG83 A0A147BA47 T1F6L5 A0A3P8TMD2 A0A3B1JWY7 A0A3B4CCU3 A0A1S3JQ95 A0A1A8M9H1 A0A0E9X7D4 A0A1A8ATX8 R4WDL1 A0A1W4Z8H0 F7FPY5 W8C601 A0A0K8UZ82 A0A034VPQ5 A0A3B4XFD9 A0A3Q2P008 A0A3Q2DSM1 A0A3B3YZY1
A0A3S2LUH6 A0A194R276 A0A194Q5Z2 A0A2P8XRL7 A0A0T6B152 A0A226EWT5 A0A182SJZ9 A0A182YBE0 A0A182PTS0 A0A182M1X8 A0A182Q4M7 A0A182NR76 A0A182VVT4 A0A182TVD0 A0A182X9K7 A0A182L8T5 Q7PUB7 A0A182RNJ7 A0A182HKJ0 A0A1D2NGH8 A0A182V4U4 A0A023EHV7 A0A1Y1MQE2 T1E8K8 A0A2M3Z8U7 A0A2M4APG7 W5J3R1 A0A084VXV3 A0A182IXM0 A0A182FRK7 D2A6A8 A0A1L8E5Q5 A0A1B0GI81 A0A1Q3FG03 B0X5P8 Q16WZ0 U5ELN7 A0A2L2Y2K6 T1DSL0 A0A0P5KDK0 A0A224YIR3 A0A0K8RH12 A0A212FMJ7 A0A1E1WWE6 A0A087U3N5 A0A1J1ITS5 A0A131Z4K4 L7LY75 A0A131XIW3 A0A023FHE6 S4PL32 A0A1B6D5I3 A0A1W4X8B9 A0A1E1XUH2 A0A023FEX5 A0A2A4K3Z8 G3MNL8 A0A1S3D024 A0A2R5LNJ4 A0A1B6J3U4 E0VA94 A0A0K8TLR8 A0A1B6LN36 A0A182K4E5 A0A023NL51 A0A2R7WRQ9 R7UGT8 A0A0L8FI18 A0A293M415 A0A3M6T475 A0A1B6HV00 A0A1Z5KUB3 A0A0K8SJF4 A7RRG4 N6UG83 A0A147BA47 T1F6L5 A0A3P8TMD2 A0A3B1JWY7 A0A3B4CCU3 A0A1S3JQ95 A0A1A8M9H1 A0A0E9X7D4 A0A1A8ATX8 R4WDL1 A0A1W4Z8H0 F7FPY5 W8C601 A0A0K8UZ82 A0A034VPQ5 A0A3B4XFD9 A0A3Q2P008 A0A3Q2DSM1 A0A3B3YZY1
PDB
6DEC
E-value=6.20696e-66,
Score=631
Ontologies
GO
PANTHER
Topology
Subcellular location
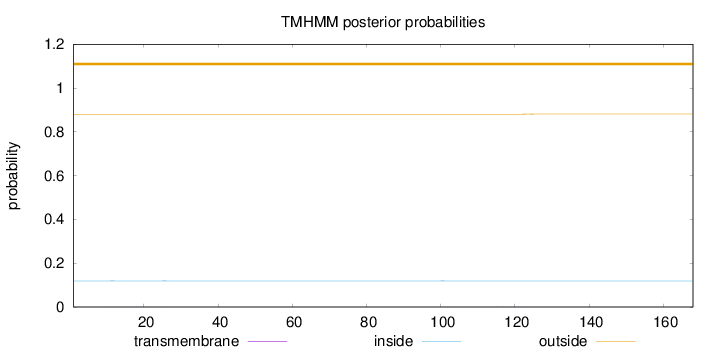
Length:
168
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01622
Exp number, first 60 AAs:
0.00295
Total prob of N-in:
0.11913
outside
1 - 168
Population Genetic Test Statistics
Pi
223.450241
Theta
143.49186
Tajima's D
1.799648
CLR
86.652069
CSRT
0.849907504624769
Interpretation
Uncertain
