Pre Gene Modal
BGIBMGA012903
Annotation
PREDICTED:_ubiquitin_fusion_degradation_protein_1_homolog_isoform_X1_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.717 Nuclear Reliability : 1.268
Sequence
CDS
ATGTTTCAATTCGGATTCAATATGTTTCACGAAATATCTAGGCCATTTAATATGACTTATAGATGCTATTCAGTCTCTATGTTACCTGGGAATGAGAGACAGGACGTTGAAAGAGGTGGTAAAATAATTATGCCCCCATCGGCATTAGAACAACTCACAAGACTCAATATAGAATATCCTATGATATTCAAGTTGACAAATAAGAAATCAAAGAGGCTCACTCATTGTGGTGTGCTAGAGTTTGTAGCTGATGAAGGAAGAGTGTATCTGCCACATTGGATGATGGCAAATCTTGTTCTAGAAGAAGGAGCTTTAATACAAATTGAAAGTGTATCACTACCTGTAGCAACATTCTCTAAGTTCCAACCTTTATCAGAAGATTTTTTGGACATTACAAATCCGAAAGCTGTATTGGAAAATTGTTTGCGTAACTTCTCATGTTTAACAACCGGTGATGTCATCGCTATAAAGTACAACTCAAAAGTCTACGAGCTGTGTGTCCTGGAGACCAAACCCGGGAATGCTGTCATCATTATAGAGTGCGATATGAATGTAGAATTTGCTCCACCAGTCGGGTATAAGGAACAAGATCACGTGTCAAGGTCAAATGATGGAGCAGTAGAAGGTATGGATGAAGATCCTGCTGCGATGATGCCCGAATCTAGTGGCTTCAGGGCGTTTAGCGGCGAAGGCAACCGCCTCGATGGAAAGAAAAAGAAGTTGATCAGCGAGAGTGATTCCGAACCGGGAACTTCGCAACCCCGCCAGTCATATGTACGAGGCATTCCGGATTACGACTTTGTGATTGGTACACTGCGATTTATCAGAAATTCAAGACCAGCCAAAGACCCTAAAGAGGAGGCTCAAACCGAACCTTTTCAAGCATTTACAGGCGAAGGATTTACATTGCGTACTGCCAAGTCTAAGAACTAA
Protein
MFQFGFNMFHEISRPFNMTYRCYSVSMLPGNERQDVERGGKIIMPPSALEQLTRLNIEYPMIFKLTNKKSKRLTHCGVLEFVADEGRVYLPHWMMANLVLEEGALIQIESVSLPVATFSKFQPLSEDFLDITNPKAVLENCLRNFSCLTTGDVIAIKYNSKVYELCVLETKPGNAVIIIECDMNVEFAPPVGYKEQDHVSRSNDGAVEGMDEDPAAMMPESSGFRAFSGEGNRLDGKKKKLISESDSEPGTSQPRQSYVRGIPDYDFVIGTLRFIRNSRPAKDPKEEAQTEPFQAFTGEGFTLRTAKSKN
Summary
Uniprot
A0A2A4K305
A0A2H1V4U7
A0A212FMJ2
H9JTP1
S4PSD6
A0A1E1WP70
+ More
A0A3S2PKI1 A0A194R2Z9 A0A194Q1F6 A0A1W4WF51 A0A2J7PMF2 A0A067R875 A0A0C9RI63 J3JWH4 A0A232F3L0 K7IRD7 E2B893 J9K1H9 C4WSF6 A0A2A3E7J7 A0A026VW06 A0A0M8ZUU9 A0A0J7KGH1 J9K462 A0A0K8RER8 A0A3L8D5S9 A0A154PE11 A0A151WSS0 A0A088A4B3 Q16WZ2 A0A023EQA0 A0A195CB84 A0A195FQP1 A0A1S3DL21 E0VMM4 Q16WZ1 A0A1S4FLI4 A0A182G4B6 A0A1E1XD09 A0A310SI08 A0A023FJT4 A0A1L8DT77 T1E370 L7M0E6 A0A131YTQ6 G3MHI1 A0A023GIB4 A0A1B0GHH7 A0A195BXD7 A0A2M4AKE4 A0A1Q3FBW4 A0A224Z338 A0A2R5L842 A0A2M4AK94 A0A182FRK6 A0A182K4E4 B0X5P5 A0A2M4BUZ0 W5J4X4 F4WXI7 A0A336M2W5 A0A2M4AKC8 A0A1B6E2B3 A0A2M4BV17 T1IT12 A0A131XGQ6 A0A182JG89 U5EN67 A0A182QHR6 A0A182PTR9 A0A182TZQ0 A0A182L8T6 Q7PUB8 A0A182HKJ1 A0A182VL45 A0A084VXV4 E2AHS1 A0A1Z5L6N8 A0A182X9K8 A0A0A9WCH1 A0A0P5RSA7 A0A0K8T3H3 A0A2L2YBJ6 A0A0P6C3X7 A0A182NR77 A0A0P4ZER3 A0A182YBD9 A0A0P5WQJ1 E9GNJ9 A0A0N8EHB2 A0A182VVT3 A0A0P5AX32 T1KMP3 A0A182RNJ8 A0A0P5Z195 A0A1B6LNQ4 A0A0A9WG80 A0A3S3NUU7 A0A182LTH9 A0A1B6HJF9
A0A3S2PKI1 A0A194R2Z9 A0A194Q1F6 A0A1W4WF51 A0A2J7PMF2 A0A067R875 A0A0C9RI63 J3JWH4 A0A232F3L0 K7IRD7 E2B893 J9K1H9 C4WSF6 A0A2A3E7J7 A0A026VW06 A0A0M8ZUU9 A0A0J7KGH1 J9K462 A0A0K8RER8 A0A3L8D5S9 A0A154PE11 A0A151WSS0 A0A088A4B3 Q16WZ2 A0A023EQA0 A0A195CB84 A0A195FQP1 A0A1S3DL21 E0VMM4 Q16WZ1 A0A1S4FLI4 A0A182G4B6 A0A1E1XD09 A0A310SI08 A0A023FJT4 A0A1L8DT77 T1E370 L7M0E6 A0A131YTQ6 G3MHI1 A0A023GIB4 A0A1B0GHH7 A0A195BXD7 A0A2M4AKE4 A0A1Q3FBW4 A0A224Z338 A0A2R5L842 A0A2M4AK94 A0A182FRK6 A0A182K4E4 B0X5P5 A0A2M4BUZ0 W5J4X4 F4WXI7 A0A336M2W5 A0A2M4AKC8 A0A1B6E2B3 A0A2M4BV17 T1IT12 A0A131XGQ6 A0A182JG89 U5EN67 A0A182QHR6 A0A182PTR9 A0A182TZQ0 A0A182L8T6 Q7PUB8 A0A182HKJ1 A0A182VL45 A0A084VXV4 E2AHS1 A0A1Z5L6N8 A0A182X9K8 A0A0A9WCH1 A0A0P5RSA7 A0A0K8T3H3 A0A2L2YBJ6 A0A0P6C3X7 A0A182NR77 A0A0P4ZER3 A0A182YBD9 A0A0P5WQJ1 E9GNJ9 A0A0N8EHB2 A0A182VVT3 A0A0P5AX32 T1KMP3 A0A182RNJ8 A0A0P5Z195 A0A1B6LNQ4 A0A0A9WG80 A0A3S3NUU7 A0A182LTH9 A0A1B6HJF9
Pubmed
22118469
19121390
23622113
26354079
24845553
22516182
+ More
28648823 20075255 20798317 24508170 30249741 17510324 24945155 20566863 26483478 28503490 24330624 25576852 26830274 22216098 28797301 20920257 23761445 21719571 28049606 20966253 12364791 14747013 17210077 24438588 28528879 25401762 26823975 26561354 25244985 21292972
28648823 20075255 20798317 24508170 30249741 17510324 24945155 20566863 26483478 28503490 24330624 25576852 26830274 22216098 28797301 20920257 23761445 21719571 28049606 20966253 12364791 14747013 17210077 24438588 28528879 25401762 26823975 26561354 25244985 21292972
EMBL
NWSH01000188
PCG78627.1
ODYU01000678
SOQ35861.1
AGBW02007651
OWR54910.1
+ More
BABH01003438 BABH01003439 GAIX01000025 JAA92535.1 GDQN01002259 JAT88795.1 RSAL01000008 RVE54020.1 KQ460878 KPJ11620.1 KQ459582 KPI98824.1 NEVH01024021 PNF17513.1 KK852677 KDR18679.1 GBYB01007945 JAG77712.1 BT127592 AEE62554.1 NNAY01001120 OXU25068.1 GL446286 EFN88095.1 ABLF02021668 AK340169 BAH70826.1 KZ288344 PBC27685.1 KK107727 EZA47978.1 KQ435840 KOX71427.1 LBMM01007874 KMQ89344.1 ABLF02031546 GADI01004222 JAA69586.1 QOIP01000013 RLU15521.1 KQ434874 KZC09654.1 KQ982769 KYQ50896.1 CH477549 EAT39129.1 GAPW01002864 JAC10734.1 KQ978023 KYM97960.1 KQ981335 KYN42617.1 DS235317 EEB14630.1 EAT39130.1 JXUM01000125 JXUM01055969 KQ561891 KQ560101 KXJ77245.1 KXJ84562.1 GFAC01002048 JAT97140.1 KQ766043 OAD53769.1 GBBK01003048 JAC21434.1 GFDF01004422 JAV09662.1 GALA01000099 JAA94753.1 GACK01008485 JAA56549.1 GEDV01006642 JAP81915.1 JO841332 AEO32949.1 GBBM01001721 JAC33697.1 AJWK01005168 KQ976401 KYM92611.1 GGFK01007881 MBW41202.1 GFDL01010043 JAV25002.1 GFPF01013091 MAA24237.1 GGLE01001550 MBY05676.1 GGFK01007831 MBW41152.1 DS232391 EDS40982.1 GGFJ01007759 MBW56900.1 ADMH02002105 ETN59006.1 GL888425 EGI61057.1 UFQT01000280 SSX22717.1 GGFK01007924 MBW41245.1 GEDC01005238 JAS32060.1 GGFJ01007758 MBW56899.1 JH431454 GEFH01003735 JAP64846.1 GANO01000716 JAB59155.1 AXCN02001469 AAAB01008987 EAA01785.3 APCN01000833 ATLV01018208 KE525224 KFB42798.1 GL439579 EFN67000.1 GFJQ02004112 JAW02858.1 GBHO01038100 GDHC01017509 JAG05504.1 JAQ01120.1 GDIQ01102512 GDIP01047873 JAL49214.1 JAM55842.1 GBRD01005757 JAG60064.1 IAAA01011635 LAA05532.1 GDIP01007354 JAM96361.1 GDIP01227050 JAI96351.1 GDIP01095616 JAM08099.1 GL732555 EFX78783.1 GDIQ01026772 JAN67965.1 GDIP01192904 JAJ30498.1 CAEY01000248 GDIP01050517 JAM53198.1 GEBQ01014679 JAT25298.1 GBHO01038101 GDHC01017188 JAG05503.1 JAQ01441.1 NCKU01004103 RWS06446.1 AXCM01001747 GECU01033427 GECU01032908 GECU01020467 GECU01018266 JAS74279.1 JAS74798.1 JAS87239.1 JAS89440.1
BABH01003438 BABH01003439 GAIX01000025 JAA92535.1 GDQN01002259 JAT88795.1 RSAL01000008 RVE54020.1 KQ460878 KPJ11620.1 KQ459582 KPI98824.1 NEVH01024021 PNF17513.1 KK852677 KDR18679.1 GBYB01007945 JAG77712.1 BT127592 AEE62554.1 NNAY01001120 OXU25068.1 GL446286 EFN88095.1 ABLF02021668 AK340169 BAH70826.1 KZ288344 PBC27685.1 KK107727 EZA47978.1 KQ435840 KOX71427.1 LBMM01007874 KMQ89344.1 ABLF02031546 GADI01004222 JAA69586.1 QOIP01000013 RLU15521.1 KQ434874 KZC09654.1 KQ982769 KYQ50896.1 CH477549 EAT39129.1 GAPW01002864 JAC10734.1 KQ978023 KYM97960.1 KQ981335 KYN42617.1 DS235317 EEB14630.1 EAT39130.1 JXUM01000125 JXUM01055969 KQ561891 KQ560101 KXJ77245.1 KXJ84562.1 GFAC01002048 JAT97140.1 KQ766043 OAD53769.1 GBBK01003048 JAC21434.1 GFDF01004422 JAV09662.1 GALA01000099 JAA94753.1 GACK01008485 JAA56549.1 GEDV01006642 JAP81915.1 JO841332 AEO32949.1 GBBM01001721 JAC33697.1 AJWK01005168 KQ976401 KYM92611.1 GGFK01007881 MBW41202.1 GFDL01010043 JAV25002.1 GFPF01013091 MAA24237.1 GGLE01001550 MBY05676.1 GGFK01007831 MBW41152.1 DS232391 EDS40982.1 GGFJ01007759 MBW56900.1 ADMH02002105 ETN59006.1 GL888425 EGI61057.1 UFQT01000280 SSX22717.1 GGFK01007924 MBW41245.1 GEDC01005238 JAS32060.1 GGFJ01007758 MBW56899.1 JH431454 GEFH01003735 JAP64846.1 GANO01000716 JAB59155.1 AXCN02001469 AAAB01008987 EAA01785.3 APCN01000833 ATLV01018208 KE525224 KFB42798.1 GL439579 EFN67000.1 GFJQ02004112 JAW02858.1 GBHO01038100 GDHC01017509 JAG05504.1 JAQ01120.1 GDIQ01102512 GDIP01047873 JAL49214.1 JAM55842.1 GBRD01005757 JAG60064.1 IAAA01011635 LAA05532.1 GDIP01007354 JAM96361.1 GDIP01227050 JAI96351.1 GDIP01095616 JAM08099.1 GL732555 EFX78783.1 GDIQ01026772 JAN67965.1 GDIP01192904 JAJ30498.1 CAEY01000248 GDIP01050517 JAM53198.1 GEBQ01014679 JAT25298.1 GBHO01038101 GDHC01017188 JAG05503.1 JAQ01441.1 NCKU01004103 RWS06446.1 AXCM01001747 GECU01033427 GECU01032908 GECU01020467 GECU01018266 JAS74279.1 JAS74798.1 JAS87239.1 JAS89440.1
Proteomes
UP000218220
UP000007151
UP000005204
UP000283053
UP000053240
UP000053268
+ More
UP000192223 UP000235965 UP000027135 UP000215335 UP000002358 UP000008237 UP000007819 UP000242457 UP000053097 UP000053105 UP000036403 UP000279307 UP000076502 UP000075809 UP000005203 UP000008820 UP000078542 UP000078541 UP000079169 UP000009046 UP000069940 UP000249989 UP000092461 UP000078540 UP000069272 UP000075881 UP000002320 UP000000673 UP000007755 UP000075880 UP000075886 UP000075885 UP000075902 UP000075882 UP000007062 UP000075840 UP000075903 UP000030765 UP000000311 UP000076407 UP000075884 UP000076408 UP000000305 UP000075920 UP000015104 UP000075900 UP000285301 UP000075883
UP000192223 UP000235965 UP000027135 UP000215335 UP000002358 UP000008237 UP000007819 UP000242457 UP000053097 UP000053105 UP000036403 UP000279307 UP000076502 UP000075809 UP000005203 UP000008820 UP000078542 UP000078541 UP000079169 UP000009046 UP000069940 UP000249989 UP000092461 UP000078540 UP000069272 UP000075881 UP000002320 UP000000673 UP000007755 UP000075880 UP000075886 UP000075885 UP000075902 UP000075882 UP000007062 UP000075840 UP000075903 UP000030765 UP000000311 UP000076407 UP000075884 UP000076408 UP000000305 UP000075920 UP000015104 UP000075900 UP000285301 UP000075883
PRIDE
Interpro
SUPFAM
SSF55298
SSF55298
Gene 3D
ProteinModelPortal
A0A2A4K305
A0A2H1V4U7
A0A212FMJ2
H9JTP1
S4PSD6
A0A1E1WP70
+ More
A0A3S2PKI1 A0A194R2Z9 A0A194Q1F6 A0A1W4WF51 A0A2J7PMF2 A0A067R875 A0A0C9RI63 J3JWH4 A0A232F3L0 K7IRD7 E2B893 J9K1H9 C4WSF6 A0A2A3E7J7 A0A026VW06 A0A0M8ZUU9 A0A0J7KGH1 J9K462 A0A0K8RER8 A0A3L8D5S9 A0A154PE11 A0A151WSS0 A0A088A4B3 Q16WZ2 A0A023EQA0 A0A195CB84 A0A195FQP1 A0A1S3DL21 E0VMM4 Q16WZ1 A0A1S4FLI4 A0A182G4B6 A0A1E1XD09 A0A310SI08 A0A023FJT4 A0A1L8DT77 T1E370 L7M0E6 A0A131YTQ6 G3MHI1 A0A023GIB4 A0A1B0GHH7 A0A195BXD7 A0A2M4AKE4 A0A1Q3FBW4 A0A224Z338 A0A2R5L842 A0A2M4AK94 A0A182FRK6 A0A182K4E4 B0X5P5 A0A2M4BUZ0 W5J4X4 F4WXI7 A0A336M2W5 A0A2M4AKC8 A0A1B6E2B3 A0A2M4BV17 T1IT12 A0A131XGQ6 A0A182JG89 U5EN67 A0A182QHR6 A0A182PTR9 A0A182TZQ0 A0A182L8T6 Q7PUB8 A0A182HKJ1 A0A182VL45 A0A084VXV4 E2AHS1 A0A1Z5L6N8 A0A182X9K8 A0A0A9WCH1 A0A0P5RSA7 A0A0K8T3H3 A0A2L2YBJ6 A0A0P6C3X7 A0A182NR77 A0A0P4ZER3 A0A182YBD9 A0A0P5WQJ1 E9GNJ9 A0A0N8EHB2 A0A182VVT3 A0A0P5AX32 T1KMP3 A0A182RNJ8 A0A0P5Z195 A0A1B6LNQ4 A0A0A9WG80 A0A3S3NUU7 A0A182LTH9 A0A1B6HJF9
A0A3S2PKI1 A0A194R2Z9 A0A194Q1F6 A0A1W4WF51 A0A2J7PMF2 A0A067R875 A0A0C9RI63 J3JWH4 A0A232F3L0 K7IRD7 E2B893 J9K1H9 C4WSF6 A0A2A3E7J7 A0A026VW06 A0A0M8ZUU9 A0A0J7KGH1 J9K462 A0A0K8RER8 A0A3L8D5S9 A0A154PE11 A0A151WSS0 A0A088A4B3 Q16WZ2 A0A023EQA0 A0A195CB84 A0A195FQP1 A0A1S3DL21 E0VMM4 Q16WZ1 A0A1S4FLI4 A0A182G4B6 A0A1E1XD09 A0A310SI08 A0A023FJT4 A0A1L8DT77 T1E370 L7M0E6 A0A131YTQ6 G3MHI1 A0A023GIB4 A0A1B0GHH7 A0A195BXD7 A0A2M4AKE4 A0A1Q3FBW4 A0A224Z338 A0A2R5L842 A0A2M4AK94 A0A182FRK6 A0A182K4E4 B0X5P5 A0A2M4BUZ0 W5J4X4 F4WXI7 A0A336M2W5 A0A2M4AKC8 A0A1B6E2B3 A0A2M4BV17 T1IT12 A0A131XGQ6 A0A182JG89 U5EN67 A0A182QHR6 A0A182PTR9 A0A182TZQ0 A0A182L8T6 Q7PUB8 A0A182HKJ1 A0A182VL45 A0A084VXV4 E2AHS1 A0A1Z5L6N8 A0A182X9K8 A0A0A9WCH1 A0A0P5RSA7 A0A0K8T3H3 A0A2L2YBJ6 A0A0P6C3X7 A0A182NR77 A0A0P4ZER3 A0A182YBD9 A0A0P5WQJ1 E9GNJ9 A0A0N8EHB2 A0A182VVT3 A0A0P5AX32 T1KMP3 A0A182RNJ8 A0A0P5Z195 A0A1B6LNQ4 A0A0A9WG80 A0A3S3NUU7 A0A182LTH9 A0A1B6HJF9
PDB
2YUJ
E-value=7.40777e-76,
Score=721
Ontologies
GO
PANTHER
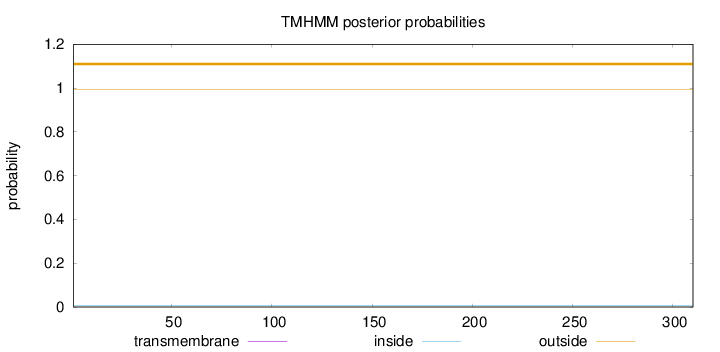
Topology
Length:
310
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000980000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00539
outside
1 - 310
Population Genetic Test Statistics
Pi
173.605862
Theta
164.180641
Tajima's D
0.378956
CLR
0.188283
CSRT
0.475226238688066
Interpretation
Uncertain
