Pre Gene Modal
BGIBMGA012936
Annotation
PREDICTED:_zinc_finger_protein_593_homolog_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.583
Sequence
CDS
ATGACATATAAGCGCAAGAAATATCATTGTGGCGACACACATTTAAAAAAGCGGTGGAGAGTGCGTAACAGAAAGAAAGATTTGGATGAAATAGACCAAGATTTAAAGGAAGAAAATGCTGAAAAATTACTGAACCAAAAAGTTGATTTAGATCTGCCAGGTGCAGCACAACATTACTGTCTACATTGTGCAAGGTATTTCATTGATGAACAAGCTTTGAATGATCATTTCAAGACAAAAGTCCATAAACGGAGATTAAAAGCACTCGAGCTTGAACCTTATACTATAGAAGAGTCAGAGAGAGCTGCTGGCCACGGGAGTTTCAAAACAGCAACAAAACGAAAAATTGTTTCACAAAATATTGAGAAATCTCAAACAGATGAAGATGGTGATGCTATTTTAGAGGAAAGTCCAAACAAAAAGAAGAAAATTGATGACAATGATATATAA
Protein
MTYKRKKYHCGDTHLKKRWRVRNRKKDLDEIDQDLKEENAEKLLNQKVDLDLPGAAQHYCLHCARYFIDEQALNDHFKTKVHKRRLKALELEPYTIEESERAAGHGSFKTATKRKIVSQNIEKSQTDEDGDAILEESPNKKKKIDDNDI
Summary
Uniprot
H9JTS4
A0A194PZR7
A0A194R1Q4
A0A0L7L3S4
A0A2H1VTP7
A0A3S2NLL5
+ More
A0A212FMD7 A0A2Z5U7Q5 A0A1W4XC83 A0A026WXA2 D2A3H6 A0A087ZU54 A0A0N0BHK7 E2BF02 E2B068 A0A2A3EH23 A0A0L7R678 A0A310SIT5 A0A158NQE7 A0A0P4VV84 F4WHA3 T1HSH2 A0A151XDM4 A0A195EWE7 A0A0A9WPC8 A0A195DUT9 A0A1Y1NJW3 A0A195CRH7 K7ILY2 E9IDI0 E9HEE7 J3JWU7 A0A0P5Y8T0 A0A0P5FH53 A0A0P4XEK8 J9K5I9 D3PFQ3 D3PJT2 A0A1L8DG91 A0A1B0D6I5 A0A232F7J8 A0A1B0CTR6 A0A210QWK8 C1BQ26 A0A0P5Y991 W8C313 T1PFR5 A0A1I8NB63 A0A2M4AV84 T1JB80 E0VBG2 A0A1S3DJZ9 B4H335 D0QWF7 B4JMY5 Q29JJ2 A0A182IX72 A0A0K8TQY1 A0A3B0JZL5 W5JL38 A0A182F8E2 A0A182YEU8 A0A2M3ZBA5 B4M7G5 A0A182MVF0 A0A2M4C244 B4L5X2 A0A1I8PR39 A0A182R8D0 B3MZ59 A0A182W2Z2 A0A2P8YPC6 B3NXU0 A0A1B0BDK3 B4PYU0 A0A023EIM9 A0A1W4VE79 T1DK94 A0A1L8EE05 K1REP2 A0A182WVF7 A0A182VCL3 A0A182U443 A0A182KXY8 Q5TT40 A0A182I936 A0A1Z5L4X7 A0A1A9YMG9 B4NE69 A0A0L0CC84 A0A3R7SQ34 A0A182PTT2 U5EIH7 A0A1A9ZKU6 A0A1A9UW44 A0A0M4EVY2 A0A182T181 A0A1S3KHE7 A0A182JYB6 A0A2C9JLB2
A0A212FMD7 A0A2Z5U7Q5 A0A1W4XC83 A0A026WXA2 D2A3H6 A0A087ZU54 A0A0N0BHK7 E2BF02 E2B068 A0A2A3EH23 A0A0L7R678 A0A310SIT5 A0A158NQE7 A0A0P4VV84 F4WHA3 T1HSH2 A0A151XDM4 A0A195EWE7 A0A0A9WPC8 A0A195DUT9 A0A1Y1NJW3 A0A195CRH7 K7ILY2 E9IDI0 E9HEE7 J3JWU7 A0A0P5Y8T0 A0A0P5FH53 A0A0P4XEK8 J9K5I9 D3PFQ3 D3PJT2 A0A1L8DG91 A0A1B0D6I5 A0A232F7J8 A0A1B0CTR6 A0A210QWK8 C1BQ26 A0A0P5Y991 W8C313 T1PFR5 A0A1I8NB63 A0A2M4AV84 T1JB80 E0VBG2 A0A1S3DJZ9 B4H335 D0QWF7 B4JMY5 Q29JJ2 A0A182IX72 A0A0K8TQY1 A0A3B0JZL5 W5JL38 A0A182F8E2 A0A182YEU8 A0A2M3ZBA5 B4M7G5 A0A182MVF0 A0A2M4C244 B4L5X2 A0A1I8PR39 A0A182R8D0 B3MZ59 A0A182W2Z2 A0A2P8YPC6 B3NXU0 A0A1B0BDK3 B4PYU0 A0A023EIM9 A0A1W4VE79 T1DK94 A0A1L8EE05 K1REP2 A0A182WVF7 A0A182VCL3 A0A182U443 A0A182KXY8 Q5TT40 A0A182I936 A0A1Z5L4X7 A0A1A9YMG9 B4NE69 A0A0L0CC84 A0A3R7SQ34 A0A182PTT2 U5EIH7 A0A1A9ZKU6 A0A1A9UW44 A0A0M4EVY2 A0A182T181 A0A1S3KHE7 A0A182JYB6 A0A2C9JLB2
Pubmed
19121390
26354079
26227816
22118469
24508170
30249741
+ More
18362917 19820115 20798317 21347285 21719571 25401762 26823975 28004739 20075255 21282665 21292972 22516182 28648823 28812685 24495485 25315136 20566863 17994087 19859648 15632085 26369729 20920257 23761445 25244985 18057021 29403074 17550304 24945155 22992520 20966253 12364791 14747013 17210077 28528879 26108605 15562597
18362917 19820115 20798317 21347285 21719571 25401762 26823975 28004739 20075255 21282665 21292972 22516182 28648823 28812685 24495485 25315136 20566863 17994087 19859648 15632085 26369729 20920257 23761445 25244985 18057021 29403074 17550304 24945155 22992520 20966253 12364791 14747013 17210077 28528879 26108605 15562597
EMBL
BABH01003440
KQ459582
KPI98822.1
KQ460878
KPJ11622.1
JTDY01003207
+ More
KOB69944.1 ODYU01004373 SOQ44187.1 RSAL01000008 RVE54022.1 AGBW02007651 OWR54912.1 AP017474 BBB06701.1 KK107078 QOIP01000011 EZA60376.1 RLU16735.1 KQ971338 EFA02318.1 KQ435750 KOX76368.1 GL447903 EFN85725.1 GL444408 EFN60925.1 KZ288260 PBC30456.1 KQ414648 KOC66281.1 KQ762237 OAD55990.1 ADTU01023180 GDRN01125025 JAI56692.1 GL888156 EGI66428.1 ACPB03008832 KQ982273 KYQ58449.1 KQ981948 KYN32558.1 GBHO01036914 GBRD01001154 GBRD01001152 GDHC01014145 JAG06690.1 JAG64667.1 JAQ04484.1 KQ980322 KYN16638.1 GEZM01001079 JAV98181.1 KQ977444 KYN02714.1 GL762454 EFZ21309.1 GL732629 EFX69887.1 BT127715 AEE62677.1 GDIP01061269 JAM42446.1 GDIQ01257434 JAJ94290.1 GDIP01243828 JAI79573.1 ABLF02034158 BT120459 ADD24099.1 BT121888 ADD38818.1 GFDF01008606 JAV05478.1 AJVK01003616 NNAY01000803 OXU26429.1 AJWK01027899 NEDP02001496 OWF53159.1 BT076705 ACO11129.1 GDIP01061270 JAM42445.1 GAMC01002932 JAC03624.1 KA647657 AFP62286.1 GGFK01011301 MBW44622.1 JH432010 DS235030 EEB10718.1 CH479205 EDW30728.1 FJ821027 ACN94661.1 CH916371 EDV92078.1 CH379063 EAL32309.1 GDAI01000834 JAI16769.1 OUUW01000003 SPP78806.1 ADMH02001252 ETN63474.1 GGFM01004977 MBW25728.1 CH940653 EDW62732.1 AXCM01000016 GGFJ01010080 MBW59221.1 CH933811 EDW06581.1 KRG07027.1 CH902632 EDV32903.1 PYGN01000453 PSN46092.1 CH954180 EDV47391.1 JXJN01012507 CM000162 EDX01026.1 GAPW01005134 JAC08464.1 GAMD01001061 JAB00530.1 GFDG01001925 JAV16874.1 JH817914 EKC39850.1 AAAB01008888 EAA08789.5 EAU76933.2 APCN01003167 GFJQ02004650 JAW02320.1 CH964239 EDW82038.1 JRES01000586 KNC30033.1 QCYY01002431 ROT70409.1 GANO01002635 JAB57236.1 CP012528 ALC49510.1
KOB69944.1 ODYU01004373 SOQ44187.1 RSAL01000008 RVE54022.1 AGBW02007651 OWR54912.1 AP017474 BBB06701.1 KK107078 QOIP01000011 EZA60376.1 RLU16735.1 KQ971338 EFA02318.1 KQ435750 KOX76368.1 GL447903 EFN85725.1 GL444408 EFN60925.1 KZ288260 PBC30456.1 KQ414648 KOC66281.1 KQ762237 OAD55990.1 ADTU01023180 GDRN01125025 JAI56692.1 GL888156 EGI66428.1 ACPB03008832 KQ982273 KYQ58449.1 KQ981948 KYN32558.1 GBHO01036914 GBRD01001154 GBRD01001152 GDHC01014145 JAG06690.1 JAG64667.1 JAQ04484.1 KQ980322 KYN16638.1 GEZM01001079 JAV98181.1 KQ977444 KYN02714.1 GL762454 EFZ21309.1 GL732629 EFX69887.1 BT127715 AEE62677.1 GDIP01061269 JAM42446.1 GDIQ01257434 JAJ94290.1 GDIP01243828 JAI79573.1 ABLF02034158 BT120459 ADD24099.1 BT121888 ADD38818.1 GFDF01008606 JAV05478.1 AJVK01003616 NNAY01000803 OXU26429.1 AJWK01027899 NEDP02001496 OWF53159.1 BT076705 ACO11129.1 GDIP01061270 JAM42445.1 GAMC01002932 JAC03624.1 KA647657 AFP62286.1 GGFK01011301 MBW44622.1 JH432010 DS235030 EEB10718.1 CH479205 EDW30728.1 FJ821027 ACN94661.1 CH916371 EDV92078.1 CH379063 EAL32309.1 GDAI01000834 JAI16769.1 OUUW01000003 SPP78806.1 ADMH02001252 ETN63474.1 GGFM01004977 MBW25728.1 CH940653 EDW62732.1 AXCM01000016 GGFJ01010080 MBW59221.1 CH933811 EDW06581.1 KRG07027.1 CH902632 EDV32903.1 PYGN01000453 PSN46092.1 CH954180 EDV47391.1 JXJN01012507 CM000162 EDX01026.1 GAPW01005134 JAC08464.1 GAMD01001061 JAB00530.1 GFDG01001925 JAV16874.1 JH817914 EKC39850.1 AAAB01008888 EAA08789.5 EAU76933.2 APCN01003167 GFJQ02004650 JAW02320.1 CH964239 EDW82038.1 JRES01000586 KNC30033.1 QCYY01002431 ROT70409.1 GANO01002635 JAB57236.1 CP012528 ALC49510.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000037510
UP000283053
UP000007151
+ More
UP000192223 UP000053097 UP000279307 UP000007266 UP000005203 UP000053105 UP000008237 UP000000311 UP000242457 UP000053825 UP000005205 UP000007755 UP000015103 UP000075809 UP000078541 UP000078492 UP000078542 UP000002358 UP000000305 UP000007819 UP000092462 UP000215335 UP000092461 UP000242188 UP000095301 UP000009046 UP000079169 UP000008744 UP000001070 UP000001819 UP000075880 UP000268350 UP000000673 UP000069272 UP000076408 UP000008792 UP000075883 UP000009192 UP000095300 UP000075900 UP000007801 UP000075920 UP000245037 UP000008711 UP000092460 UP000002282 UP000192221 UP000005408 UP000076407 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840 UP000092443 UP000007798 UP000037069 UP000283509 UP000075885 UP000092445 UP000078200 UP000092553 UP000075901 UP000085678 UP000075881 UP000076420
UP000192223 UP000053097 UP000279307 UP000007266 UP000005203 UP000053105 UP000008237 UP000000311 UP000242457 UP000053825 UP000005205 UP000007755 UP000015103 UP000075809 UP000078541 UP000078492 UP000078542 UP000002358 UP000000305 UP000007819 UP000092462 UP000215335 UP000092461 UP000242188 UP000095301 UP000009046 UP000079169 UP000008744 UP000001070 UP000001819 UP000075880 UP000268350 UP000000673 UP000069272 UP000076408 UP000008792 UP000075883 UP000009192 UP000095300 UP000075900 UP000007801 UP000075920 UP000245037 UP000008711 UP000092460 UP000002282 UP000192221 UP000005408 UP000076407 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840 UP000092443 UP000007798 UP000037069 UP000283509 UP000075885 UP000092445 UP000078200 UP000092553 UP000075901 UP000085678 UP000075881 UP000076420
PRIDE
Interpro
ProteinModelPortal
H9JTS4
A0A194PZR7
A0A194R1Q4
A0A0L7L3S4
A0A2H1VTP7
A0A3S2NLL5
+ More
A0A212FMD7 A0A2Z5U7Q5 A0A1W4XC83 A0A026WXA2 D2A3H6 A0A087ZU54 A0A0N0BHK7 E2BF02 E2B068 A0A2A3EH23 A0A0L7R678 A0A310SIT5 A0A158NQE7 A0A0P4VV84 F4WHA3 T1HSH2 A0A151XDM4 A0A195EWE7 A0A0A9WPC8 A0A195DUT9 A0A1Y1NJW3 A0A195CRH7 K7ILY2 E9IDI0 E9HEE7 J3JWU7 A0A0P5Y8T0 A0A0P5FH53 A0A0P4XEK8 J9K5I9 D3PFQ3 D3PJT2 A0A1L8DG91 A0A1B0D6I5 A0A232F7J8 A0A1B0CTR6 A0A210QWK8 C1BQ26 A0A0P5Y991 W8C313 T1PFR5 A0A1I8NB63 A0A2M4AV84 T1JB80 E0VBG2 A0A1S3DJZ9 B4H335 D0QWF7 B4JMY5 Q29JJ2 A0A182IX72 A0A0K8TQY1 A0A3B0JZL5 W5JL38 A0A182F8E2 A0A182YEU8 A0A2M3ZBA5 B4M7G5 A0A182MVF0 A0A2M4C244 B4L5X2 A0A1I8PR39 A0A182R8D0 B3MZ59 A0A182W2Z2 A0A2P8YPC6 B3NXU0 A0A1B0BDK3 B4PYU0 A0A023EIM9 A0A1W4VE79 T1DK94 A0A1L8EE05 K1REP2 A0A182WVF7 A0A182VCL3 A0A182U443 A0A182KXY8 Q5TT40 A0A182I936 A0A1Z5L4X7 A0A1A9YMG9 B4NE69 A0A0L0CC84 A0A3R7SQ34 A0A182PTT2 U5EIH7 A0A1A9ZKU6 A0A1A9UW44 A0A0M4EVY2 A0A182T181 A0A1S3KHE7 A0A182JYB6 A0A2C9JLB2
A0A212FMD7 A0A2Z5U7Q5 A0A1W4XC83 A0A026WXA2 D2A3H6 A0A087ZU54 A0A0N0BHK7 E2BF02 E2B068 A0A2A3EH23 A0A0L7R678 A0A310SIT5 A0A158NQE7 A0A0P4VV84 F4WHA3 T1HSH2 A0A151XDM4 A0A195EWE7 A0A0A9WPC8 A0A195DUT9 A0A1Y1NJW3 A0A195CRH7 K7ILY2 E9IDI0 E9HEE7 J3JWU7 A0A0P5Y8T0 A0A0P5FH53 A0A0P4XEK8 J9K5I9 D3PFQ3 D3PJT2 A0A1L8DG91 A0A1B0D6I5 A0A232F7J8 A0A1B0CTR6 A0A210QWK8 C1BQ26 A0A0P5Y991 W8C313 T1PFR5 A0A1I8NB63 A0A2M4AV84 T1JB80 E0VBG2 A0A1S3DJZ9 B4H335 D0QWF7 B4JMY5 Q29JJ2 A0A182IX72 A0A0K8TQY1 A0A3B0JZL5 W5JL38 A0A182F8E2 A0A182YEU8 A0A2M3ZBA5 B4M7G5 A0A182MVF0 A0A2M4C244 B4L5X2 A0A1I8PR39 A0A182R8D0 B3MZ59 A0A182W2Z2 A0A2P8YPC6 B3NXU0 A0A1B0BDK3 B4PYU0 A0A023EIM9 A0A1W4VE79 T1DK94 A0A1L8EE05 K1REP2 A0A182WVF7 A0A182VCL3 A0A182U443 A0A182KXY8 Q5TT40 A0A182I936 A0A1Z5L4X7 A0A1A9YMG9 B4NE69 A0A0L0CC84 A0A3R7SQ34 A0A182PTT2 U5EIH7 A0A1A9ZKU6 A0A1A9UW44 A0A0M4EVY2 A0A182T181 A0A1S3KHE7 A0A182JYB6 A0A2C9JLB2
PDB
1ZR9
E-value=1.4928e-17,
Score=213
Ontologies
GO
PANTHER
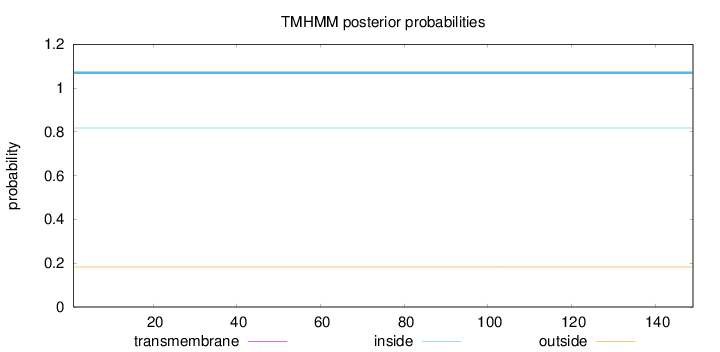
Topology
Length:
149
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.81752
inside
1 - 149
Population Genetic Test Statistics
Pi
345.011081
Theta
217.174419
Tajima's D
1.761558
CLR
0.132963
CSRT
0.846307684615769
Interpretation
Uncertain
