Gene
KWMTBOMO09745
Pre Gene Modal
BGIBMGA012902
Annotation
PREDICTED:_transport_and_Golgi_organization_protein_2_[Bombyx_mori]
Full name
Transport and Golgi organization protein 2
Location in the cell
Cytoplasmic Reliability : 1.82 Nuclear Reliability : 1.502
Sequence
CDS
ATGTGTATATTATTCGTATACAATGGAAGCTATGATGCAGACAGTGACTACAGTCTGATAGTGGCAACAAATAGAGATGAATTTTATGATCGACCTTCTGCAGAACTTGCGCCTTGGAAAGACAATCCAAATATCATTGGAGGATTAGATTTAGAAGGGGGCTGTGAAGGAGGGACGTGGCTTGCTGTGTCTCCCCTCAGAAAGAAAATGGGAGTACTTTTGAATTTACCAAATACACCAAAGAAAAATGCAAAAAGCCGAGGTAAAATTGTAGCAGAATTTGTGAAAAGTAAAAATGAGGCTGCATCATATGTTCAAGAAATGAAATCTTATTTTGAAGAATGTAATAATTTCATTTTTGTAGCAATGGACTTTGGAAACACCACGCCAGTCATAAATAGCTTCACGAATGTCACAAACTCAATAACAACACATACAGATCCTTGTTTGGGTTTCGGAAATAGTTTACCCGATATGCCATTGAAAAAAGTTGAAGCCGGCCTTACTAAAATGCATGATATTTGTAAGGGACTCAATAAAATTTCTATGAAAAGCAAACTCTTAGAAGAATTGACTGCACTCTTGAAGTGTAATGAAAGGCATCTACCTGACGCCCAACTACAAGAACGTAGGCCTAATTTATATGAAGAATTAAGTTCTATATTTGTTTGTGTACCTGAAGAAAGATATGGAACGAGGGCGCAAACTATATTGCTTCTCACGAAAACTGGTCATTTAGAAGTACACGAAATATCCATGAAATCACCAATTGATAAAATAGAACCCAAGTGGGAAAAACATAAATACCAATTTGACCTGTGA
Protein
MCILFVYNGSYDADSDYSLIVATNRDEFYDRPSAELAPWKDNPNIIGGLDLEGGCEGGTWLAVSPLRKKMGVLLNLPNTPKKNAKSRGKIVAEFVKSKNEAASYVQEMKSYFEECNNFIFVAMDFGNTTPVINSFTNVTNSITTHTDPCLGFGNSLPDMPLKKVEAGLTKMHDICKGLNKISMKSKLLEELTALLKCNERHLPDAQLQERRPNLYEELSSIFVCVPEERYGTRAQTILLLTKTGHLEVHEISMKSPIDKIEPKWEKHKYQFDL
Summary
Description
May play a role in Golgi organization.
Similarity
Belongs to the Tango2 family.
Keywords
Alternative splicing
Complete proteome
Cytoplasm
Golgi apparatus
Reference proteome
Feature
chain Transport and Golgi organization protein 2
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JTP0
A0A2A4K4N7
A0A2A4K2Y7
A0A212FMF3
A0A3S2M8Q7
A0A194Q5Y7
+ More
A0A2H1VTT3 A0A2W1C104 A0A0L7L366 A0A194R281 A0A2Z5UIL8 A0A1I8NS59 U5ETR2 A0A1I8N4S6 A0A067R2Z1 D2A5G9 A0A182SVA3 A0A0J7NAU1 A0A1Y1MF26 A0A1Q3FHE6 A0A1S3JXG9 A0A182K672 A0A182RNB4 A0A182MBA0 W8AQE4 A0A0M5JDV2 Q16NI8 A0A182VYK0 A0A1L8DW97 A0A182P6N0 A0A154PJF6 E2AQB6 K7ITV2 A0A182UM18 W5JFZ7 A0A182WRR0 A0A182FPG7 A0A232F0N0 A0A1I8NS63 A0A1L8DWA9 A0A0K8WEX5 Q7QD53 A0A182TS50 B4M1B8 E0VH46 A0A026WDA6 A0A182LG08 A0A182HU18 A0A0R3P290 A0A182Q2I1 J9JTQ4 A0A182XWC6 A0A3B0KDX8 A0A1Z5KWY8 A0A084WP18 A0A2A3E9K0 A0A147BL27 A0A224YDW3 T1J2R1 A0A131YMH6 A0A0N7ZDD6 A0A443SGC0 B4JL84 V5I119 A0A0Q5TKS2 A0A0P9C159 N6UJV1 A0A0R1EAM1 M9PEI3 Q9VYA8 A0A023FJD9 B4L3C0 G3MQ42 A0A023GHH1 A0A182NDW9 B7PW18 A0A182JE38 A0A1W4VH83 A0A131XQA7 V4ACJ8 A0A1E1X7P0 B4NDC7 A0A2S2R1D5 K1R1M6 T1K1C7 A0A0L7RFK1 A0A087TNF6 L7MLR4 Q9U1Q8 A0A401SE81 K6YZG2 E3N0I8 A0A260ZS46 U3I7K4 A0A151NS40
A0A2H1VTT3 A0A2W1C104 A0A0L7L366 A0A194R281 A0A2Z5UIL8 A0A1I8NS59 U5ETR2 A0A1I8N4S6 A0A067R2Z1 D2A5G9 A0A182SVA3 A0A0J7NAU1 A0A1Y1MF26 A0A1Q3FHE6 A0A1S3JXG9 A0A182K672 A0A182RNB4 A0A182MBA0 W8AQE4 A0A0M5JDV2 Q16NI8 A0A182VYK0 A0A1L8DW97 A0A182P6N0 A0A154PJF6 E2AQB6 K7ITV2 A0A182UM18 W5JFZ7 A0A182WRR0 A0A182FPG7 A0A232F0N0 A0A1I8NS63 A0A1L8DWA9 A0A0K8WEX5 Q7QD53 A0A182TS50 B4M1B8 E0VH46 A0A026WDA6 A0A182LG08 A0A182HU18 A0A0R3P290 A0A182Q2I1 J9JTQ4 A0A182XWC6 A0A3B0KDX8 A0A1Z5KWY8 A0A084WP18 A0A2A3E9K0 A0A147BL27 A0A224YDW3 T1J2R1 A0A131YMH6 A0A0N7ZDD6 A0A443SGC0 B4JL84 V5I119 A0A0Q5TKS2 A0A0P9C159 N6UJV1 A0A0R1EAM1 M9PEI3 Q9VYA8 A0A023FJD9 B4L3C0 G3MQ42 A0A023GHH1 A0A182NDW9 B7PW18 A0A182JE38 A0A1W4VH83 A0A131XQA7 V4ACJ8 A0A1E1X7P0 B4NDC7 A0A2S2R1D5 K1R1M6 T1K1C7 A0A0L7RFK1 A0A087TNF6 L7MLR4 Q9U1Q8 A0A401SE81 K6YZG2 E3N0I8 A0A260ZS46 U3I7K4 A0A151NS40
Pubmed
19121390
22118469
26354079
28756777
26227816
25315136
+ More
24845553 18362917 19820115 28004739 24495485 17510324 20798317 20075255 20920257 23761445 28648823 12364791 14747013 17210077 17994087 20566863 24508170 20966253 15632085 25244985 28528879 24438588 29652888 28797301 26830274 25765539 23537049 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 16452979 22216098 28049606 23254933 28503490 22992520 25576852 9851916 30297745 25009843 23749191 22293439
24845553 18362917 19820115 28004739 24495485 17510324 20798317 20075255 20920257 23761445 28648823 12364791 14747013 17210077 17994087 20566863 24508170 20966253 15632085 25244985 28528879 24438588 29652888 28797301 26830274 25765539 23537049 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 16452979 22216098 28049606 23254933 28503490 22992520 25576852 9851916 30297745 25009843 23749191 22293439
EMBL
BABH01003440
NWSH01000188
PCG78620.1
PCG78621.1
AGBW02007651
OWR54913.1
+ More
RSAL01000008 RVE54023.1 KQ459582 KPI98820.1 ODYU01004373 SOQ44188.1 KZ149898 PZC78646.1 JTDY01003207 KOB69948.1 KQ460878 KPJ11624.1 AP017474 BBB06702.1 GANO01001788 JAB58083.1 KK853042 KDR12151.1 KQ971345 EFA05078.1 LBMM01007445 KMQ89725.1 GEZM01037814 JAV81917.1 GFDL01008081 JAV26964.1 AXCM01005292 GAMC01018273 JAB88282.1 CP012528 ALC49494.1 CH477823 EAT35904.1 EAT35905.1 EAT35906.1 GFDF01003380 JAV10704.1 KQ434924 KZC11604.1 GL441721 EFN64374.1 ADMH02001267 ETN63297.1 NNAY01001370 OXU24211.1 GFDF01003381 JAV10703.1 GDHF01002924 JAI49390.1 AAAB01008859 EAA07544.4 CH940651 EDW65472.1 DS235158 EEB12702.1 KK107260 EZA54060.1 APCN01000599 CH379066 KRT07320.1 AXCN02001589 ABLF02006773 OUUW01000011 SPP86500.1 GFJQ02007425 JAV99544.1 ATLV01024815 KE525361 KFB51962.1 KZ288327 PBC27962.1 GEGO01003897 JAR91507.1 GFPF01004752 MAA15898.1 JH431806 GEDV01008772 JAP79785.1 GDRN01043973 JAI66952.1 NCKV01002656 RWS26559.1 CH916370 EDW00337.1 GANP01003495 JAB80973.1 CH954180 KQS30378.1 CH902640 KPU77355.1 APGK01021186 KB740293 KB631672 ENN80941.1 ERL85268.1 CM000162 KRK06239.1 AE014298 AGB95367.1 AY069312 BT089002 GBBK01003203 JAC21279.1 CH933810 EDW07048.1 JO843993 AEO35610.1 GBBM01002007 JAC33411.1 ABJB010013628 ABJB010345996 ABJB010692915 ABJB010915330 ABJB011022104 ABJB011129956 DS804579 EEC10790.1 GEFH01000206 JAP68375.1 KB200406 ESP01734.1 GFAC01003903 JAT95285.1 CH964239 EDW82836.2 GGMS01014593 MBY83796.1 JH816018 EKC35040.1 CAEY01001351 KQ414599 KOC69757.1 KK116039 KFM66645.1 GACK01000242 JAA64792.1 BX284605 CAB60443.1 BEZZ01000216 GCC28717.1 BAEP01000023 GAC23557.1 DS268505 EFP13367.1 NMWX01000050 OZF88550.1 ADON01104759 ADON01104760 AKHW03002185 KYO39520.1
RSAL01000008 RVE54023.1 KQ459582 KPI98820.1 ODYU01004373 SOQ44188.1 KZ149898 PZC78646.1 JTDY01003207 KOB69948.1 KQ460878 KPJ11624.1 AP017474 BBB06702.1 GANO01001788 JAB58083.1 KK853042 KDR12151.1 KQ971345 EFA05078.1 LBMM01007445 KMQ89725.1 GEZM01037814 JAV81917.1 GFDL01008081 JAV26964.1 AXCM01005292 GAMC01018273 JAB88282.1 CP012528 ALC49494.1 CH477823 EAT35904.1 EAT35905.1 EAT35906.1 GFDF01003380 JAV10704.1 KQ434924 KZC11604.1 GL441721 EFN64374.1 ADMH02001267 ETN63297.1 NNAY01001370 OXU24211.1 GFDF01003381 JAV10703.1 GDHF01002924 JAI49390.1 AAAB01008859 EAA07544.4 CH940651 EDW65472.1 DS235158 EEB12702.1 KK107260 EZA54060.1 APCN01000599 CH379066 KRT07320.1 AXCN02001589 ABLF02006773 OUUW01000011 SPP86500.1 GFJQ02007425 JAV99544.1 ATLV01024815 KE525361 KFB51962.1 KZ288327 PBC27962.1 GEGO01003897 JAR91507.1 GFPF01004752 MAA15898.1 JH431806 GEDV01008772 JAP79785.1 GDRN01043973 JAI66952.1 NCKV01002656 RWS26559.1 CH916370 EDW00337.1 GANP01003495 JAB80973.1 CH954180 KQS30378.1 CH902640 KPU77355.1 APGK01021186 KB740293 KB631672 ENN80941.1 ERL85268.1 CM000162 KRK06239.1 AE014298 AGB95367.1 AY069312 BT089002 GBBK01003203 JAC21279.1 CH933810 EDW07048.1 JO843993 AEO35610.1 GBBM01002007 JAC33411.1 ABJB010013628 ABJB010345996 ABJB010692915 ABJB010915330 ABJB011022104 ABJB011129956 DS804579 EEC10790.1 GEFH01000206 JAP68375.1 KB200406 ESP01734.1 GFAC01003903 JAT95285.1 CH964239 EDW82836.2 GGMS01014593 MBY83796.1 JH816018 EKC35040.1 CAEY01001351 KQ414599 KOC69757.1 KK116039 KFM66645.1 GACK01000242 JAA64792.1 BX284605 CAB60443.1 BEZZ01000216 GCC28717.1 BAEP01000023 GAC23557.1 DS268505 EFP13367.1 NMWX01000050 OZF88550.1 ADON01104759 ADON01104760 AKHW03002185 KYO39520.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053268
UP000037510
+ More
UP000053240 UP000095300 UP000095301 UP000027135 UP000007266 UP000075901 UP000036403 UP000085678 UP000075881 UP000075900 UP000075883 UP000092553 UP000008820 UP000075920 UP000075885 UP000076502 UP000000311 UP000002358 UP000075903 UP000000673 UP000076407 UP000069272 UP000215335 UP000007062 UP000075902 UP000008792 UP000009046 UP000053097 UP000075882 UP000075840 UP000001819 UP000075886 UP000007819 UP000076408 UP000268350 UP000030765 UP000242457 UP000288716 UP000001070 UP000008711 UP000007801 UP000019118 UP000030742 UP000002282 UP000000803 UP000009192 UP000075884 UP000001555 UP000075880 UP000192221 UP000030746 UP000007798 UP000005408 UP000015104 UP000053825 UP000054359 UP000001940 UP000287033 UP000006263 UP000008281 UP000216624 UP000016666 UP000050525
UP000053240 UP000095300 UP000095301 UP000027135 UP000007266 UP000075901 UP000036403 UP000085678 UP000075881 UP000075900 UP000075883 UP000092553 UP000008820 UP000075920 UP000075885 UP000076502 UP000000311 UP000002358 UP000075903 UP000000673 UP000076407 UP000069272 UP000215335 UP000007062 UP000075902 UP000008792 UP000009046 UP000053097 UP000075882 UP000075840 UP000001819 UP000075886 UP000007819 UP000076408 UP000268350 UP000030765 UP000242457 UP000288716 UP000001070 UP000008711 UP000007801 UP000019118 UP000030742 UP000002282 UP000000803 UP000009192 UP000075884 UP000001555 UP000075880 UP000192221 UP000030746 UP000007798 UP000005408 UP000015104 UP000053825 UP000054359 UP000001940 UP000287033 UP000006263 UP000008281 UP000216624 UP000016666 UP000050525
Interpro
SUPFAM
SSF57667
SSF57667
ProteinModelPortal
H9JTP0
A0A2A4K4N7
A0A2A4K2Y7
A0A212FMF3
A0A3S2M8Q7
A0A194Q5Y7
+ More
A0A2H1VTT3 A0A2W1C104 A0A0L7L366 A0A194R281 A0A2Z5UIL8 A0A1I8NS59 U5ETR2 A0A1I8N4S6 A0A067R2Z1 D2A5G9 A0A182SVA3 A0A0J7NAU1 A0A1Y1MF26 A0A1Q3FHE6 A0A1S3JXG9 A0A182K672 A0A182RNB4 A0A182MBA0 W8AQE4 A0A0M5JDV2 Q16NI8 A0A182VYK0 A0A1L8DW97 A0A182P6N0 A0A154PJF6 E2AQB6 K7ITV2 A0A182UM18 W5JFZ7 A0A182WRR0 A0A182FPG7 A0A232F0N0 A0A1I8NS63 A0A1L8DWA9 A0A0K8WEX5 Q7QD53 A0A182TS50 B4M1B8 E0VH46 A0A026WDA6 A0A182LG08 A0A182HU18 A0A0R3P290 A0A182Q2I1 J9JTQ4 A0A182XWC6 A0A3B0KDX8 A0A1Z5KWY8 A0A084WP18 A0A2A3E9K0 A0A147BL27 A0A224YDW3 T1J2R1 A0A131YMH6 A0A0N7ZDD6 A0A443SGC0 B4JL84 V5I119 A0A0Q5TKS2 A0A0P9C159 N6UJV1 A0A0R1EAM1 M9PEI3 Q9VYA8 A0A023FJD9 B4L3C0 G3MQ42 A0A023GHH1 A0A182NDW9 B7PW18 A0A182JE38 A0A1W4VH83 A0A131XQA7 V4ACJ8 A0A1E1X7P0 B4NDC7 A0A2S2R1D5 K1R1M6 T1K1C7 A0A0L7RFK1 A0A087TNF6 L7MLR4 Q9U1Q8 A0A401SE81 K6YZG2 E3N0I8 A0A260ZS46 U3I7K4 A0A151NS40
A0A2H1VTT3 A0A2W1C104 A0A0L7L366 A0A194R281 A0A2Z5UIL8 A0A1I8NS59 U5ETR2 A0A1I8N4S6 A0A067R2Z1 D2A5G9 A0A182SVA3 A0A0J7NAU1 A0A1Y1MF26 A0A1Q3FHE6 A0A1S3JXG9 A0A182K672 A0A182RNB4 A0A182MBA0 W8AQE4 A0A0M5JDV2 Q16NI8 A0A182VYK0 A0A1L8DW97 A0A182P6N0 A0A154PJF6 E2AQB6 K7ITV2 A0A182UM18 W5JFZ7 A0A182WRR0 A0A182FPG7 A0A232F0N0 A0A1I8NS63 A0A1L8DWA9 A0A0K8WEX5 Q7QD53 A0A182TS50 B4M1B8 E0VH46 A0A026WDA6 A0A182LG08 A0A182HU18 A0A0R3P290 A0A182Q2I1 J9JTQ4 A0A182XWC6 A0A3B0KDX8 A0A1Z5KWY8 A0A084WP18 A0A2A3E9K0 A0A147BL27 A0A224YDW3 T1J2R1 A0A131YMH6 A0A0N7ZDD6 A0A443SGC0 B4JL84 V5I119 A0A0Q5TKS2 A0A0P9C159 N6UJV1 A0A0R1EAM1 M9PEI3 Q9VYA8 A0A023FJD9 B4L3C0 G3MQ42 A0A023GHH1 A0A182NDW9 B7PW18 A0A182JE38 A0A1W4VH83 A0A131XQA7 V4ACJ8 A0A1E1X7P0 B4NDC7 A0A2S2R1D5 K1R1M6 T1K1C7 A0A0L7RFK1 A0A087TNF6 L7MLR4 Q9U1Q8 A0A401SE81 K6YZG2 E3N0I8 A0A260ZS46 U3I7K4 A0A151NS40
Ontologies
PANTHER
Topology
Subcellular location
Cytoplasm
Golgi apparatus
Golgi apparatus
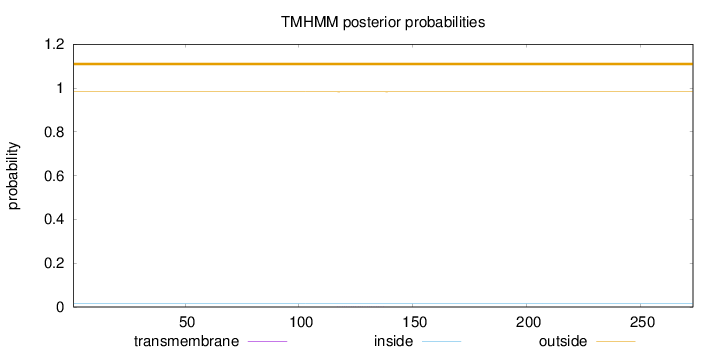
Length:
273
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00693
Exp number, first 60 AAs:
0.00033
Total prob of N-in:
0.01714
outside
1 - 273
Population Genetic Test Statistics
Pi
193.798441
Theta
168.268259
Tajima's D
0.743489
CLR
0.658177
CSRT
0.587820608969552
Interpretation
Uncertain
