Gene
KWMTBOMO09743
Pre Gene Modal
BGIBMGA012901
Annotation
FancJ-like_protein_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.41
Sequence
CDS
ATGGGTACACACGGAGCTACCAGCATAGATGAAGAACAATCCCAAACCCGAAAGAAATACAAAGCTGAGAACATTACCAATATATCTGACTCTCCACGGTCTCCGTCTAACTCACAGAACCCGCCGAAACATGAACGGGTTCCAGTCGTGTACTACGGCACCAGAACCCACAAACAGCTACAGCAGGTGATCAAAGAGTTCAAACGCACGGTCTACTGCAAGGAGATACGTATGTCGATACTGGCCGGCAGAGACCGGACCTGTCTAATGCCATTCGATCGTAAAATCTGGAAAACCAAGGACGATATGTGCGCTGAGTGCATCAAAACCAAATCAAGTCTGATGAATACGTCATCACCGCAAACGTCAAGCTGCAAATTCTACGACAACCGAATGTCGTTGACGCACAGCAACTTGCCCCCGGCCTTCGACATCGAGGACTTGGTCGAAGCCGGAGCGAAACTGAAGGCCTGCCCTTACTTCGCTGCCAAGAGCATGGCCGCAAAAGCAAAAATCATCTTCTGTCCTTACAACTATCTGATCAACCCTTTCATTAGGGAGCGGATGAGGATCAATTTAGCAGACAACATAGTCGTTATAGACGAAGGGCACAACATCGAAGACATCTGCAGAGACGAGGCCACCTGCACCATCGAGAGGAGACGGATATCTGAAGCTGTTGATCAACTGCAGTACGCCAAGAGGTACGTTAAGATGGGCCAGGATGAGGCTGCAAGAGGGGAAACAATAGATTTTTTCATCAAGACATTACAAATATGGGATAAGTGTCTAATAGAACAATCCGATCGCGTATCGCGCGAGAACACCGACATGTGGAACATGAACGAGTTCATGGCTATGCTAAATGAAAATGGCATCGGACAAAACAATTATCATAATTTCAATTTTCATTTATCGAAGCTGTTACGTATGCAAGATGATTTGTACGGTGTGGGACAACACACGATCGCCTTGTTGGAAACAATGGACTCCGCTCTCGGGTACCTTTTCAAGGACGAATGTAAACACGCCGAAGACTTCGTGCCGATGTTGCTGAAGGGTGACGACTCGCGCGCGCTCTATCGGCGACGGTCGTATACCGGCGAGTTCAAAGGTGCGAACATATCGATACGTCTTCTATGCATGAGTCCGGCGGTGCTGTTCGAGGGTCTGACGAAGGCTAGATCTATAATACTGGCCTCCGGTACACTGTCGCCCGTGGGCAGCATGAAGCACGAGCTCGGCGCCGAGTTCAACGTGTCGTGTTTGAACCACGTCATCTCTAGAGAACGGGTATGGATAGGAACGTTAAGAAACTGCGAGAGCTCGGAACAATTGAAATGTACGAAGGATAATATGGAGAAGAAACAAGTACAGGATTCCATAGGGAAGGCTATATTGAGGGTGTGTGACGTCACGCCCCATGGAGTGCTGTGCTTTCATTCCTCGTACACATCCATGAGGTTGCTACATCAAAGATGGCAACAAACTGGCTTGTGGGACAAATTGAATAGCCTAAAATATATATTTAAGGAGAGCGACAATACGACTGATCATGACGAAATTATGAAGGAGTACTACAGCGCCGCGGAGGTGGATCGAGGCGCGATCCTGTTCGCCGTGTACAGGGGGAAGGTCTCCGAGGGCATGGACTTCAGGGACCGGCAAGCCAGGGCCGTTGTGACGATCGGCGTGCCCTTCCCAACGATCGGAGACAAGATGGTCACGGAGAAAATGAAATACAACGACAAGCATTCGAATGGGAAGAAGTTAATGCGCGGCTGGGACTGGTACAACGTGCAAGCCTTCAGAGCCATGAACCAAGCCATAGGACGTTGCGTGAGGCACCTCAACGACTGGGGCGCCGTTCTGATGATCGACGAGAGAATCGCAGAACAAAGGAATCTTGAATATCTATCTAAATGGGTGCGAGACTTCCTCGGAAACAACCATTTTACTTTCGATGAATTAATGCATAGACAAAACAGCCTCACAAGATTTATGGAGAACATGAGCAAAGAATAA
Protein
MGTHGATSIDEEQSQTRKKYKAENITNISDSPRSPSNSQNPPKHERVPVVYYGTRTHKQLQQVIKEFKRTVYCKEIRMSILAGRDRTCLMPFDRKIWKTKDDMCAECIKTKSSLMNTSSPQTSSCKFYDNRMSLTHSNLPPAFDIEDLVEAGAKLKACPYFAAKSMAAKAKIIFCPYNYLINPFIRERMRINLADNIVVIDEGHNIEDICRDEATCTIERRRISEAVDQLQYAKRYVKMGQDEAARGETIDFFIKTLQIWDKCLIEQSDRVSRENTDMWNMNEFMAMLNENGIGQNNYHNFNFHLSKLLRMQDDLYGVGQHTIALLETMDSALGYLFKDECKHAEDFVPMLLKGDDSRALYRRRSYTGEFKGANISIRLLCMSPAVLFEGLTKARSIILASGTLSPVGSMKHELGAEFNVSCLNHVISRERVWIGTLRNCESSEQLKCTKDNMEKKQVQDSIGKAILRVCDVTPHGVLCFHSSYTSMRLLHQRWQQTGLWDKLNSLKYIFKESDNTTDHDEIMKEYYSAAEVDRGAILFAVYRGKVSEGMDFRDRQARAVVTIGVPFPTIGDKMVTEKMKYNDKHSNGKKLMRGWDWYNVQAFRAMNQAIGRCVRHLNDWGAVLMIDERIAEQRNLEYLSKWVRDFLGNNHFTFDELMHRQNSLTRFMENMSKE
Summary
Uniprot
A9EEZ8
A0A194PZT2
A0A437BUI9
A0A2W1BYG9
A0A2H1VVE1
A0A212FME2
+ More
A0A3Q1IQZ1 H2T3N5 A0A293LE89 A0A3P9I1H1 A0A3P9I1E8 A0A3P9JFQ7 A0A3B4VLU2 A0A3B3H8F0 H2MIU7 A0A3B3HB80 A0A3B4ZN16 A0A3B3HKH5 A0A3B3H805 A0A3P9M0B9 A0A3P8XN58 A0A3M6UJH1 I3JWB2 A0A3B4HCA1 A0A3Q2V3Q1 A0A151WPQ7 H3DA69 F6ULH9 A0A3P9AWL3 A0A3P8QPV4 A0A3B3BW47 A0A3Q3L5G2 A0A146ZNM1 A0A3Q2Q5U0 A0A146VAX1 A0A1S3NRR4 A0A3B3VGS8 A0A3B3BVC5 E2AJR2 A0A3P9PLY0 A0A3B3DDW3 A0A087YEW9 A0A3P9PLJ7 V4AHF8 H0ZBR6 A0A3Q2Q5D1 A0A093R258 A0A1X7UE18 A0A3P8S870 A0A3Q2NMX5 A0A0P7UJ65 A0A0A0ABS1 A0A2R8QEM7 A0A091HX30 A0A091M9K4 A0A0Q3PY07 B7ZDD5 A0A093CZN1 A0A094LMH0 A0A2R8QLG8 A0A1A7XTS7 A0A2R8Q596 A0A3B5LA20 A0A2R8QLM4 A0A2R8QCP4 A0A2R8S0B7 A8QLH7 A0A3Q3K0C3 A0A2G8KDW1 A0A3B5RC02 A0A3Q2XFD4 A0A091UHS8 A0A3Q1C674 M3ZU62 A0A1L8HG08 A0A093KLT1 A0A2I0LYG7 K1QY39 A0A1V4K1Y6 A0A3Q0RSW1 G3I6L7 A0A2Y9RA33 A0A091L764 A0A099ZPJ6 A0A2U9B3Y3 A0A2D0SWT7 A0A091IUC2 G1N6L5 A0A2I4D943 A0A1W4ZSQ4 A0A3B4DJZ9 A0A091UGH3 A0A091SND1 A0A3Q2ZJI7 A0A093ETD5 A0A091QBS8 A0A3Q1ER48 A0A3Q1H996
A0A3Q1IQZ1 H2T3N5 A0A293LE89 A0A3P9I1H1 A0A3P9I1E8 A0A3P9JFQ7 A0A3B4VLU2 A0A3B3H8F0 H2MIU7 A0A3B3HB80 A0A3B4ZN16 A0A3B3HKH5 A0A3B3H805 A0A3P9M0B9 A0A3P8XN58 A0A3M6UJH1 I3JWB2 A0A3B4HCA1 A0A3Q2V3Q1 A0A151WPQ7 H3DA69 F6ULH9 A0A3P9AWL3 A0A3P8QPV4 A0A3B3BW47 A0A3Q3L5G2 A0A146ZNM1 A0A3Q2Q5U0 A0A146VAX1 A0A1S3NRR4 A0A3B3VGS8 A0A3B3BVC5 E2AJR2 A0A3P9PLY0 A0A3B3DDW3 A0A087YEW9 A0A3P9PLJ7 V4AHF8 H0ZBR6 A0A3Q2Q5D1 A0A093R258 A0A1X7UE18 A0A3P8S870 A0A3Q2NMX5 A0A0P7UJ65 A0A0A0ABS1 A0A2R8QEM7 A0A091HX30 A0A091M9K4 A0A0Q3PY07 B7ZDD5 A0A093CZN1 A0A094LMH0 A0A2R8QLG8 A0A1A7XTS7 A0A2R8Q596 A0A3B5LA20 A0A2R8QLM4 A0A2R8QCP4 A0A2R8S0B7 A8QLH7 A0A3Q3K0C3 A0A2G8KDW1 A0A3B5RC02 A0A3Q2XFD4 A0A091UHS8 A0A3Q1C674 M3ZU62 A0A1L8HG08 A0A093KLT1 A0A2I0LYG7 K1QY39 A0A1V4K1Y6 A0A3Q0RSW1 G3I6L7 A0A2Y9RA33 A0A091L764 A0A099ZPJ6 A0A2U9B3Y3 A0A2D0SWT7 A0A091IUC2 G1N6L5 A0A2I4D943 A0A1W4ZSQ4 A0A3B4DJZ9 A0A091UGH3 A0A091SND1 A0A3Q2ZJI7 A0A093ETD5 A0A091QBS8 A0A3Q1ER48 A0A3Q1H996
Pubmed
EMBL
AB369600
BAF94023.1
KQ459582
KPI98837.1
RSAL01000008
RVE54025.1
+ More
KZ149898 PZC78644.1 ODYU01004372 SOQ44184.1 AGBW02007651 OWR54915.1 GFWV01002313 MAA27043.1 RCHS01001413 RMX53830.1 AERX01031533 AERX01031534 AERX01031535 AERX01031536 AERX01031537 KQ982860 KYQ49788.1 AAMC01027091 AAMC01027092 AAMC01027093 AAMC01027094 AAMC01027095 AAMC01027096 AAMC01027097 AAMC01027098 GCES01018410 JAR67913.1 GCES01071882 JAR14441.1 GL440062 EFN66346.1 AYCK01010880 AYCK01010881 AYCK01010882 AYCK01010883 KB201794 ESO94630.1 ABQF01026546 ABQF01026547 KL444839 KFW95163.1 JARO02001813 KPP74356.1 KL871238 KGL91372.1 AL954187 AL954322 BX005270 CT573367 KL217847 KFO99724.1 KK523406 KFP67239.1 LMAW01002498 KQK80948.1 KL467871 KFV17727.1 KL360316 KFZ64989.1 HADW01020033 SBP21433.1 AL954326 EF088194 ABO27623.1 MRZV01000662 PIK46155.1 KK436363 KFQ90339.1 CM004468 OCT94971.1 KK588553 KFV98625.1 AKCR02000061 PKK22472.1 JH817109 EKC26471.1 LSYS01005108 OPJ78490.1 JH001376 EGV98907.1 KL306641 KFP52284.1 KL895544 KGL82705.1 CP026245 AWO98448.1 KK501002 KFP11907.1 KK450974 KFQ73583.1 KK477535 KFQ59455.1 KK371737 KFV43893.1 KK692032 KFQ22059.1
KZ149898 PZC78644.1 ODYU01004372 SOQ44184.1 AGBW02007651 OWR54915.1 GFWV01002313 MAA27043.1 RCHS01001413 RMX53830.1 AERX01031533 AERX01031534 AERX01031535 AERX01031536 AERX01031537 KQ982860 KYQ49788.1 AAMC01027091 AAMC01027092 AAMC01027093 AAMC01027094 AAMC01027095 AAMC01027096 AAMC01027097 AAMC01027098 GCES01018410 JAR67913.1 GCES01071882 JAR14441.1 GL440062 EFN66346.1 AYCK01010880 AYCK01010881 AYCK01010882 AYCK01010883 KB201794 ESO94630.1 ABQF01026546 ABQF01026547 KL444839 KFW95163.1 JARO02001813 KPP74356.1 KL871238 KGL91372.1 AL954187 AL954322 BX005270 CT573367 KL217847 KFO99724.1 KK523406 KFP67239.1 LMAW01002498 KQK80948.1 KL467871 KFV17727.1 KL360316 KFZ64989.1 HADW01020033 SBP21433.1 AL954326 EF088194 ABO27623.1 MRZV01000662 PIK46155.1 KK436363 KFQ90339.1 CM004468 OCT94971.1 KK588553 KFV98625.1 AKCR02000061 PKK22472.1 JH817109 EKC26471.1 LSYS01005108 OPJ78490.1 JH001376 EGV98907.1 KL306641 KFP52284.1 KL895544 KGL82705.1 CP026245 AWO98448.1 KK501002 KFP11907.1 KK450974 KFQ73583.1 KK477535 KFQ59455.1 KK371737 KFV43893.1 KK692032 KFQ22059.1
Proteomes
UP000053268
UP000283053
UP000007151
UP000265040
UP000005226
UP000265200
+ More
UP000261420 UP000001038 UP000261400 UP000265180 UP000265140 UP000275408 UP000005207 UP000261460 UP000264840 UP000075809 UP000007303 UP000008143 UP000265160 UP000265100 UP000261560 UP000261640 UP000265000 UP000087266 UP000261500 UP000000311 UP000242638 UP000028760 UP000030746 UP000007754 UP000007879 UP000265080 UP000034805 UP000053858 UP000000437 UP000054308 UP000051836 UP000261380 UP000261600 UP000230750 UP000002852 UP000264820 UP000257160 UP000186698 UP000053872 UP000005408 UP000190648 UP000261340 UP000001075 UP000248480 UP000053641 UP000246464 UP000221080 UP000053119 UP000001645 UP000192220 UP000192224 UP000261440 UP000264800 UP000257200
UP000261420 UP000001038 UP000261400 UP000265180 UP000265140 UP000275408 UP000005207 UP000261460 UP000264840 UP000075809 UP000007303 UP000008143 UP000265160 UP000265100 UP000261560 UP000261640 UP000265000 UP000087266 UP000261500 UP000000311 UP000242638 UP000028760 UP000030746 UP000007754 UP000007879 UP000265080 UP000034805 UP000053858 UP000000437 UP000054308 UP000051836 UP000261380 UP000261600 UP000230750 UP000002852 UP000264820 UP000257160 UP000186698 UP000053872 UP000005408 UP000190648 UP000261340 UP000001075 UP000248480 UP000053641 UP000246464 UP000221080 UP000053119 UP000001645 UP000192220 UP000192224 UP000261440 UP000264800 UP000257200
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A9EEZ8
A0A194PZT2
A0A437BUI9
A0A2W1BYG9
A0A2H1VVE1
A0A212FME2
+ More
A0A3Q1IQZ1 H2T3N5 A0A293LE89 A0A3P9I1H1 A0A3P9I1E8 A0A3P9JFQ7 A0A3B4VLU2 A0A3B3H8F0 H2MIU7 A0A3B3HB80 A0A3B4ZN16 A0A3B3HKH5 A0A3B3H805 A0A3P9M0B9 A0A3P8XN58 A0A3M6UJH1 I3JWB2 A0A3B4HCA1 A0A3Q2V3Q1 A0A151WPQ7 H3DA69 F6ULH9 A0A3P9AWL3 A0A3P8QPV4 A0A3B3BW47 A0A3Q3L5G2 A0A146ZNM1 A0A3Q2Q5U0 A0A146VAX1 A0A1S3NRR4 A0A3B3VGS8 A0A3B3BVC5 E2AJR2 A0A3P9PLY0 A0A3B3DDW3 A0A087YEW9 A0A3P9PLJ7 V4AHF8 H0ZBR6 A0A3Q2Q5D1 A0A093R258 A0A1X7UE18 A0A3P8S870 A0A3Q2NMX5 A0A0P7UJ65 A0A0A0ABS1 A0A2R8QEM7 A0A091HX30 A0A091M9K4 A0A0Q3PY07 B7ZDD5 A0A093CZN1 A0A094LMH0 A0A2R8QLG8 A0A1A7XTS7 A0A2R8Q596 A0A3B5LA20 A0A2R8QLM4 A0A2R8QCP4 A0A2R8S0B7 A8QLH7 A0A3Q3K0C3 A0A2G8KDW1 A0A3B5RC02 A0A3Q2XFD4 A0A091UHS8 A0A3Q1C674 M3ZU62 A0A1L8HG08 A0A093KLT1 A0A2I0LYG7 K1QY39 A0A1V4K1Y6 A0A3Q0RSW1 G3I6L7 A0A2Y9RA33 A0A091L764 A0A099ZPJ6 A0A2U9B3Y3 A0A2D0SWT7 A0A091IUC2 G1N6L5 A0A2I4D943 A0A1W4ZSQ4 A0A3B4DJZ9 A0A091UGH3 A0A091SND1 A0A3Q2ZJI7 A0A093ETD5 A0A091QBS8 A0A3Q1ER48 A0A3Q1H996
A0A3Q1IQZ1 H2T3N5 A0A293LE89 A0A3P9I1H1 A0A3P9I1E8 A0A3P9JFQ7 A0A3B4VLU2 A0A3B3H8F0 H2MIU7 A0A3B3HB80 A0A3B4ZN16 A0A3B3HKH5 A0A3B3H805 A0A3P9M0B9 A0A3P8XN58 A0A3M6UJH1 I3JWB2 A0A3B4HCA1 A0A3Q2V3Q1 A0A151WPQ7 H3DA69 F6ULH9 A0A3P9AWL3 A0A3P8QPV4 A0A3B3BW47 A0A3Q3L5G2 A0A146ZNM1 A0A3Q2Q5U0 A0A146VAX1 A0A1S3NRR4 A0A3B3VGS8 A0A3B3BVC5 E2AJR2 A0A3P9PLY0 A0A3B3DDW3 A0A087YEW9 A0A3P9PLJ7 V4AHF8 H0ZBR6 A0A3Q2Q5D1 A0A093R258 A0A1X7UE18 A0A3P8S870 A0A3Q2NMX5 A0A0P7UJ65 A0A0A0ABS1 A0A2R8QEM7 A0A091HX30 A0A091M9K4 A0A0Q3PY07 B7ZDD5 A0A093CZN1 A0A094LMH0 A0A2R8QLG8 A0A1A7XTS7 A0A2R8Q596 A0A3B5LA20 A0A2R8QLM4 A0A2R8QCP4 A0A2R8S0B7 A8QLH7 A0A3Q3K0C3 A0A2G8KDW1 A0A3B5RC02 A0A3Q2XFD4 A0A091UHS8 A0A3Q1C674 M3ZU62 A0A1L8HG08 A0A093KLT1 A0A2I0LYG7 K1QY39 A0A1V4K1Y6 A0A3Q0RSW1 G3I6L7 A0A2Y9RA33 A0A091L764 A0A099ZPJ6 A0A2U9B3Y3 A0A2D0SWT7 A0A091IUC2 G1N6L5 A0A2I4D943 A0A1W4ZSQ4 A0A3B4DJZ9 A0A091UGH3 A0A091SND1 A0A3Q2ZJI7 A0A093ETD5 A0A091QBS8 A0A3Q1ER48 A0A3Q1H996
PDB
6NMI
E-value=1.28226e-24,
Score=283
Ontologies
PATHWAY
GO
Topology
Subcellular location
Nucleus
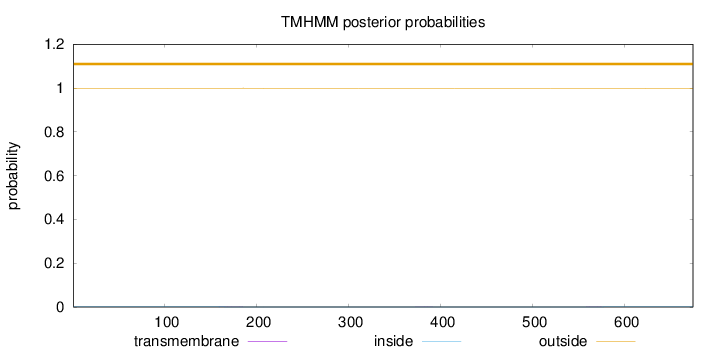
Length:
674
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01926
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00099
outside
1 - 674
Population Genetic Test Statistics
Pi
271.72534
Theta
172.254687
Tajima's D
1.59459
CLR
0.516252
CSRT
0.809009549522524
Interpretation
Uncertain
