Gene
KWMTBOMO09734 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012894
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_adenosine_deaminase_CECR1-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.43
Sequence
CDS
ATGCCAACAAAATTGACTGTGTTCTTATGGCTGAGCATCATAATGCAAGCGTTTTGCAACACAACCCACGAAACAGTAAAGAAGAAAAGAAATGAACTATTTGAAAAAGAATTGGAAATGATGCTCGGACGCGACATTGTATTGAATGAAAACGAAACTAAAGTCAACGAGATATTTATGAAACTTAAAACGAAAGAACTAGACCAAGGATTCCGGCACAGACGTCCTTTTGTTTTATCCAAACACTTCTTTGAATACAAAGATGATGTCAAGAAAACTGAACTGTACAAGCTGATTAAAAAAATGCCAAAAGGTGCGGTGCTTCATGCCCACGATACTTCACTCTTAGGTCCAGATTACGTATTAGAATTAACATACTGGAAATATTTATATGTTTGTTTTATTGACGACGAAGTATTATTTAGATTTGCGAAGACTTTACCGACCACACCGTGTGCAACAAAATGGCAGCTTGTTAAAGAAGCAAGATACTCATCGGGTAATGTTGATGAATTCGATGATCATCTCCGAAAAGAGTTCACGTTAGTCGTAGATGATTATGTAAAGAAGTACCCAGATAATAATGCCATTTGGAAGAGCTTCAAGAAGTATTTCAAAACAATTTTATCGATCGTCTCTTACAAACCGGCATGGGAGCAATACTTTTACGACGCGCTAAAGGAGTATCGTCATGACAATGTATTCTTTCTTGAAGTGAGAAGTATTTTACCAACGCTTTACGATTTAGATGGAAACGTTTATGACCCAGTAGCAACCGCTGAATCATATCAGAAAGTAACTGAACAATTCATGCATGATTATCCAGATTTTATGGGTGCGAAACTTATTTACGCGCCATCTAGAGGGACAGAGCAATTTACTTTCGATGAATATTTGAGATTCGCAAAAGAAATCAAACGTAAGCTACCGGATTTCTTTGCTGGTTTCGATCTAGTCAATCACGAGGATGCGGGACCGCCAAACAAACACTTTCTACCACAATTGGTCAATGAAAAAGATAACTTGAACTACTATTTCCATGCCGCCGAAACTAATTGGAACGGAATGGATTCAGACGAAAACCTGTATGACGCTTTGGCTCTGGATACTAAACGTATCGGTCACGGATTCGCTCTGATAAAGCATCCACTTTTAATAGATGAAGTAAGAAAGAGAGATATCGCCATAGAAGTGAATGTCATTTCCAATAATGTTCTTTGTTTAGTCAGTGATATTAGGAATCACCCGCTTTCGTCTTATTTAGCGCAAGGCTTGCCTGTCGTGGTGTCGAGTGATGATCGCGGTGCATGGGAAGCGGACCCTTTGTCCCATGACTTCTATGCCGCATTCGTTGGAGTAGCCAGCCGCCATGCAGACCTGAGGCTTCTGAAAAAGCTCGCTCTGAATTCGATACGCTACAGTACGTATTCGGACAAAAGTAAAATGTTTGCTGAATTCGAAAGCCGATGGTCACAATTCATTAATGCTGTGAAAGATAAACATGTTTAG
Protein
MPTKLTVFLWLSIIMQAFCNTTHETVKKKRNELFEKELEMMLGRDIVLNENETKVNEIFMKLKTKELDQGFRHRRPFVLSKHFFEYKDDVKKTELYKLIKKMPKGAVLHAHDTSLLGPDYVLELTYWKYLYVCFIDDEVLFRFAKTLPTTPCATKWQLVKEARYSSGNVDEFDDHLRKEFTLVVDDYVKKYPDNNAIWKSFKKYFKTILSIVSYKPAWEQYFYDALKEYRHDNVFFLEVRSILPTLYDLDGNVYDPVATAESYQKVTEQFMHDYPDFMGAKLIYAPSRGTEQFTFDEYLRFAKEIKRKLPDFFAGFDLVNHEDAGPPNKHFLPQLVNEKDNLNYYFHAAETNWNGMDSDENLYDALALDTKRIGHGFALIKHPLLIDEVRKRDIAIEVNVISNNVLCLVSDIRNHPLSSYLAQGLPVVVSSDDRGAWEADPLSHDFYAAFVGVASRHADLRLLKKLALNSIRYSTYSDKSKMFAEFESRWSQFINAVKDKHV
Summary
Uniprot
H9JTN2
A0A0L7LPB3
A0A194Q0L6
A0A2A4JD81
A0A194R1N1
A0A2W1C0J8
+ More
A6M8U8 H9JTN3 H9JTS7 A7BEX8 A0A2H1VBY9 A0A212FMK2 A0A2H1VBV3 A0A2A4JCS6 A0A2W1BUH1 A0A194Q1I1 A0A2H1VAK6 A0A194R6V8 A0A212F3M6 I4DP17 A0A0L7K4B9 A0A212FMK6 A0A194R1C8 A0A2H1W0Y1 A0A2W1BU87 H9JTS6 A0A0L0C082 A0A2A4K563 A0A2W1BWC1 A0A194PZU2 A0A2A4K5F4 Q9VVK5 B4QNZ1 B4HKW2 W5JVM2 A0A0L7LNV3 A0A0L7LUI0 A0A3G5BIM1 Q26642 A0A3B0KIU2 A0A336LUY4 Q2M0I1 B3NHR7 A0A1W4W4X4 A0A182QSP6 B4PJJ8 A0A182MQK5 A0A1A9XQJ8 A0A1I8PKD4 A0A182N9D8 B4N6J7 A0A1A9WPY1 A0A1I8MAS7 T1P9Q7 A0A1A9V4I6 A0A0K8VRD8 A0A1A9ZTP5 A0A0A1X851 A0A034VKN8 B3M4T9 A0A0M4EGX7 A0A0M4F150 A0A182GU02 A0A182WBF6 W8BGP3 A0A1S4FB76 W8B694 Q179D5 A0A182IWP5 A0A182FEG8 W5JUB6 B4KZH9 B4LEX7 A0A1B0GAX2 A0A2C9GS79 Q5TWK1 A0A182JEX0 A0A182UV11 A0A1Q3FRC2 A0A182RJI0 A0A182L3Q7 A0A1S4GU77 B4IR34 A0A182TC49 D6WEK0 A0A084WKR7 A0A182TCZ3 A0A0L7R411 A0A1Q3FNZ6 A0A0L7LPB2 B0WWE6 A0A182PIL7 A0A182K2P4 B4J3K4 A0A0P4VQR1 A0A182Y803 A0A084WKR6 A0A1W4WZ20 A0A182WWF7 A0A1W6EVV3
A6M8U8 H9JTN3 H9JTS7 A7BEX8 A0A2H1VBY9 A0A212FMK2 A0A2H1VBV3 A0A2A4JCS6 A0A2W1BUH1 A0A194Q1I1 A0A2H1VAK6 A0A194R6V8 A0A212F3M6 I4DP17 A0A0L7K4B9 A0A212FMK6 A0A194R1C8 A0A2H1W0Y1 A0A2W1BU87 H9JTS6 A0A0L0C082 A0A2A4K563 A0A2W1BWC1 A0A194PZU2 A0A2A4K5F4 Q9VVK5 B4QNZ1 B4HKW2 W5JVM2 A0A0L7LNV3 A0A0L7LUI0 A0A3G5BIM1 Q26642 A0A3B0KIU2 A0A336LUY4 Q2M0I1 B3NHR7 A0A1W4W4X4 A0A182QSP6 B4PJJ8 A0A182MQK5 A0A1A9XQJ8 A0A1I8PKD4 A0A182N9D8 B4N6J7 A0A1A9WPY1 A0A1I8MAS7 T1P9Q7 A0A1A9V4I6 A0A0K8VRD8 A0A1A9ZTP5 A0A0A1X851 A0A034VKN8 B3M4T9 A0A0M4EGX7 A0A0M4F150 A0A182GU02 A0A182WBF6 W8BGP3 A0A1S4FB76 W8B694 Q179D5 A0A182IWP5 A0A182FEG8 W5JUB6 B4KZH9 B4LEX7 A0A1B0GAX2 A0A2C9GS79 Q5TWK1 A0A182JEX0 A0A182UV11 A0A1Q3FRC2 A0A182RJI0 A0A182L3Q7 A0A1S4GU77 B4IR34 A0A182TC49 D6WEK0 A0A084WKR7 A0A182TCZ3 A0A0L7R411 A0A1Q3FNZ6 A0A0L7LPB2 B0WWE6 A0A182PIL7 A0A182K2P4 B4J3K4 A0A0P4VQR1 A0A182Y803 A0A084WKR6 A0A1W4WZ20 A0A182WWF7 A0A1W6EVV3
Pubmed
19121390
26227816
26354079
28756777
17439545
22118469
+ More
22651552 26108605 10731132 11738815 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 22936249 20920257 23761445 30400621 8662793 10756095 10967093 11562360 17011515 15632085 23185243 17550304 25315136 25830018 25348373 18057021 26483478 24495485 17510324 12364791 20966253 18362917 19820115 24438588 25244985
22651552 26108605 10731132 11738815 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 22936249 20920257 23761445 30400621 8662793 10756095 10967093 11562360 17011515 15632085 23185243 17550304 25315136 25830018 25348373 18057021 26483478 24495485 17510324 12364791 20966253 18362917 19820115 24438588 25244985
EMBL
BABH01003476
JTDY01000431
KOB77209.1
KQ459582
KPI98848.1
NWSH01001926
+ More
PCG69648.1 KQ460878 KPJ11597.1 KZ149898 PZC78640.1 DQ322663 ABC50165.1 BABH01003475 AB183871 BAF73622.1 ODYU01001720 SOQ38311.1 AGBW02007651 OWR54920.1 ODYU01001533 SOQ37882.1 PCG69649.1 PZC78639.1 KPI98849.1 SOQ37883.1 KPJ11596.1 AGBW02010531 OWR48335.1 AK403248 BAM19657.1 JTDY01010697 KOB58090.1 OWR54919.1 KPJ11598.1 ODYU01005668 SOQ46760.1 PZC78642.1 BABH01003474 JRES01001077 KNC25705.1 NWSH01000164 PCG78913.1 PZC78641.1 KPI98847.1 PCG78912.1 AE014296 AY058655 AF337554 AAF49306.1 AAL13884.1 AAL40913.1 AAS64976.1 CM000363 CM002912 EDX10893.1 KMZ00301.1 CH480815 EDW41917.1 ADMH02000347 ETN66884.1 KOB77203.1 JTDY01000099 KOB78871.1 MK075218 AYV99621.1 D83125 BAA11812.1 OUUW01000009 SPP85001.1 UFQT01000125 SSX20499.1 CH379069 EAL30951.3 KRT07791.1 CH954178 EDV51932.1 AXCN02000560 CM000159 EDW94686.1 AXCM01005713 CH964161 EDW79986.2 KA644678 AFP59307.1 GDHF01010872 JAI41442.1 GBXI01015529 GBXI01007382 JAC98762.1 JAD06910.1 GAKP01016292 GAKP01016291 GAKP01016290 JAC42662.1 CH902618 EDV40513.1 KPU78606.1 CP012525 ALC42888.1 ALC45000.1 JXUM01088455 KQ563703 KXJ73474.1 GAMC01017721 GAMC01017720 JAB88834.1 GAMC01017719 GAMC01017718 JAB88836.1 CH477354 EAT42833.1 ETN66883.1 CH933809 EDW17906.1 KRG05831.1 CH940647 EDW69146.1 KRF84202.1 CCAG010001199 APCN01003684 AAAB01008807 EAL41919.3 GFDL01005072 JAV29973.1 CH690550 EDW29854.1 KQ971318 EFA00425.1 ATLV01024134 KE525349 KFB50811.1 KQ414661 KOC65564.1 GFDL01005842 JAV29203.1 KOB77204.1 DS232144 EDS36038.1 CH916366 EDV96206.1 GDRN01107117 JAI57470.1 KFB50810.1 KY563447 ARK19856.1
PCG69648.1 KQ460878 KPJ11597.1 KZ149898 PZC78640.1 DQ322663 ABC50165.1 BABH01003475 AB183871 BAF73622.1 ODYU01001720 SOQ38311.1 AGBW02007651 OWR54920.1 ODYU01001533 SOQ37882.1 PCG69649.1 PZC78639.1 KPI98849.1 SOQ37883.1 KPJ11596.1 AGBW02010531 OWR48335.1 AK403248 BAM19657.1 JTDY01010697 KOB58090.1 OWR54919.1 KPJ11598.1 ODYU01005668 SOQ46760.1 PZC78642.1 BABH01003474 JRES01001077 KNC25705.1 NWSH01000164 PCG78913.1 PZC78641.1 KPI98847.1 PCG78912.1 AE014296 AY058655 AF337554 AAF49306.1 AAL13884.1 AAL40913.1 AAS64976.1 CM000363 CM002912 EDX10893.1 KMZ00301.1 CH480815 EDW41917.1 ADMH02000347 ETN66884.1 KOB77203.1 JTDY01000099 KOB78871.1 MK075218 AYV99621.1 D83125 BAA11812.1 OUUW01000009 SPP85001.1 UFQT01000125 SSX20499.1 CH379069 EAL30951.3 KRT07791.1 CH954178 EDV51932.1 AXCN02000560 CM000159 EDW94686.1 AXCM01005713 CH964161 EDW79986.2 KA644678 AFP59307.1 GDHF01010872 JAI41442.1 GBXI01015529 GBXI01007382 JAC98762.1 JAD06910.1 GAKP01016292 GAKP01016291 GAKP01016290 JAC42662.1 CH902618 EDV40513.1 KPU78606.1 CP012525 ALC42888.1 ALC45000.1 JXUM01088455 KQ563703 KXJ73474.1 GAMC01017721 GAMC01017720 JAB88834.1 GAMC01017719 GAMC01017718 JAB88836.1 CH477354 EAT42833.1 ETN66883.1 CH933809 EDW17906.1 KRG05831.1 CH940647 EDW69146.1 KRF84202.1 CCAG010001199 APCN01003684 AAAB01008807 EAL41919.3 GFDL01005072 JAV29973.1 CH690550 EDW29854.1 KQ971318 EFA00425.1 ATLV01024134 KE525349 KFB50811.1 KQ414661 KOC65564.1 GFDL01005842 JAV29203.1 KOB77204.1 DS232144 EDS36038.1 CH916366 EDV96206.1 GDRN01107117 JAI57470.1 KFB50810.1 KY563447 ARK19856.1
Proteomes
UP000005204
UP000037510
UP000053268
UP000218220
UP000053240
UP000007151
+ More
UP000037069 UP000000803 UP000000304 UP000001292 UP000000673 UP000268350 UP000001819 UP000008711 UP000192221 UP000075886 UP000002282 UP000075883 UP000092443 UP000095300 UP000075884 UP000007798 UP000091820 UP000095301 UP000078200 UP000092445 UP000007801 UP000092553 UP000069940 UP000249989 UP000075920 UP000008820 UP000075880 UP000069272 UP000009192 UP000008792 UP000092444 UP000075840 UP000007062 UP000075903 UP000075900 UP000075882 UP000008744 UP000075901 UP000007266 UP000030765 UP000075902 UP000053825 UP000002320 UP000075885 UP000075881 UP000001070 UP000076408 UP000192223 UP000076407
UP000037069 UP000000803 UP000000304 UP000001292 UP000000673 UP000268350 UP000001819 UP000008711 UP000192221 UP000075886 UP000002282 UP000075883 UP000092443 UP000095300 UP000075884 UP000007798 UP000091820 UP000095301 UP000078200 UP000092445 UP000007801 UP000092553 UP000069940 UP000249989 UP000075920 UP000008820 UP000075880 UP000069272 UP000009192 UP000008792 UP000092444 UP000075840 UP000007062 UP000075903 UP000075900 UP000075882 UP000008744 UP000075901 UP000007266 UP000030765 UP000075902 UP000053825 UP000002320 UP000075885 UP000075881 UP000001070 UP000076408 UP000192223 UP000076407
Interpro
SUPFAM
SSF51556
SSF51556
ProteinModelPortal
H9JTN2
A0A0L7LPB3
A0A194Q0L6
A0A2A4JD81
A0A194R1N1
A0A2W1C0J8
+ More
A6M8U8 H9JTN3 H9JTS7 A7BEX8 A0A2H1VBY9 A0A212FMK2 A0A2H1VBV3 A0A2A4JCS6 A0A2W1BUH1 A0A194Q1I1 A0A2H1VAK6 A0A194R6V8 A0A212F3M6 I4DP17 A0A0L7K4B9 A0A212FMK6 A0A194R1C8 A0A2H1W0Y1 A0A2W1BU87 H9JTS6 A0A0L0C082 A0A2A4K563 A0A2W1BWC1 A0A194PZU2 A0A2A4K5F4 Q9VVK5 B4QNZ1 B4HKW2 W5JVM2 A0A0L7LNV3 A0A0L7LUI0 A0A3G5BIM1 Q26642 A0A3B0KIU2 A0A336LUY4 Q2M0I1 B3NHR7 A0A1W4W4X4 A0A182QSP6 B4PJJ8 A0A182MQK5 A0A1A9XQJ8 A0A1I8PKD4 A0A182N9D8 B4N6J7 A0A1A9WPY1 A0A1I8MAS7 T1P9Q7 A0A1A9V4I6 A0A0K8VRD8 A0A1A9ZTP5 A0A0A1X851 A0A034VKN8 B3M4T9 A0A0M4EGX7 A0A0M4F150 A0A182GU02 A0A182WBF6 W8BGP3 A0A1S4FB76 W8B694 Q179D5 A0A182IWP5 A0A182FEG8 W5JUB6 B4KZH9 B4LEX7 A0A1B0GAX2 A0A2C9GS79 Q5TWK1 A0A182JEX0 A0A182UV11 A0A1Q3FRC2 A0A182RJI0 A0A182L3Q7 A0A1S4GU77 B4IR34 A0A182TC49 D6WEK0 A0A084WKR7 A0A182TCZ3 A0A0L7R411 A0A1Q3FNZ6 A0A0L7LPB2 B0WWE6 A0A182PIL7 A0A182K2P4 B4J3K4 A0A0P4VQR1 A0A182Y803 A0A084WKR6 A0A1W4WZ20 A0A182WWF7 A0A1W6EVV3
A6M8U8 H9JTN3 H9JTS7 A7BEX8 A0A2H1VBY9 A0A212FMK2 A0A2H1VBV3 A0A2A4JCS6 A0A2W1BUH1 A0A194Q1I1 A0A2H1VAK6 A0A194R6V8 A0A212F3M6 I4DP17 A0A0L7K4B9 A0A212FMK6 A0A194R1C8 A0A2H1W0Y1 A0A2W1BU87 H9JTS6 A0A0L0C082 A0A2A4K563 A0A2W1BWC1 A0A194PZU2 A0A2A4K5F4 Q9VVK5 B4QNZ1 B4HKW2 W5JVM2 A0A0L7LNV3 A0A0L7LUI0 A0A3G5BIM1 Q26642 A0A3B0KIU2 A0A336LUY4 Q2M0I1 B3NHR7 A0A1W4W4X4 A0A182QSP6 B4PJJ8 A0A182MQK5 A0A1A9XQJ8 A0A1I8PKD4 A0A182N9D8 B4N6J7 A0A1A9WPY1 A0A1I8MAS7 T1P9Q7 A0A1A9V4I6 A0A0K8VRD8 A0A1A9ZTP5 A0A0A1X851 A0A034VKN8 B3M4T9 A0A0M4EGX7 A0A0M4F150 A0A182GU02 A0A182WBF6 W8BGP3 A0A1S4FB76 W8B694 Q179D5 A0A182IWP5 A0A182FEG8 W5JUB6 B4KZH9 B4LEX7 A0A1B0GAX2 A0A2C9GS79 Q5TWK1 A0A182JEX0 A0A182UV11 A0A1Q3FRC2 A0A182RJI0 A0A182L3Q7 A0A1S4GU77 B4IR34 A0A182TC49 D6WEK0 A0A084WKR7 A0A182TCZ3 A0A0L7R411 A0A1Q3FNZ6 A0A0L7LPB2 B0WWE6 A0A182PIL7 A0A182K2P4 B4J3K4 A0A0P4VQR1 A0A182Y803 A0A084WKR6 A0A1W4WZ20 A0A182WWF7 A0A1W6EVV3
PDB
3LGG
E-value=1.0672e-95,
Score=894
Ontologies
KEGG
PATHWAY
GO
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.965369
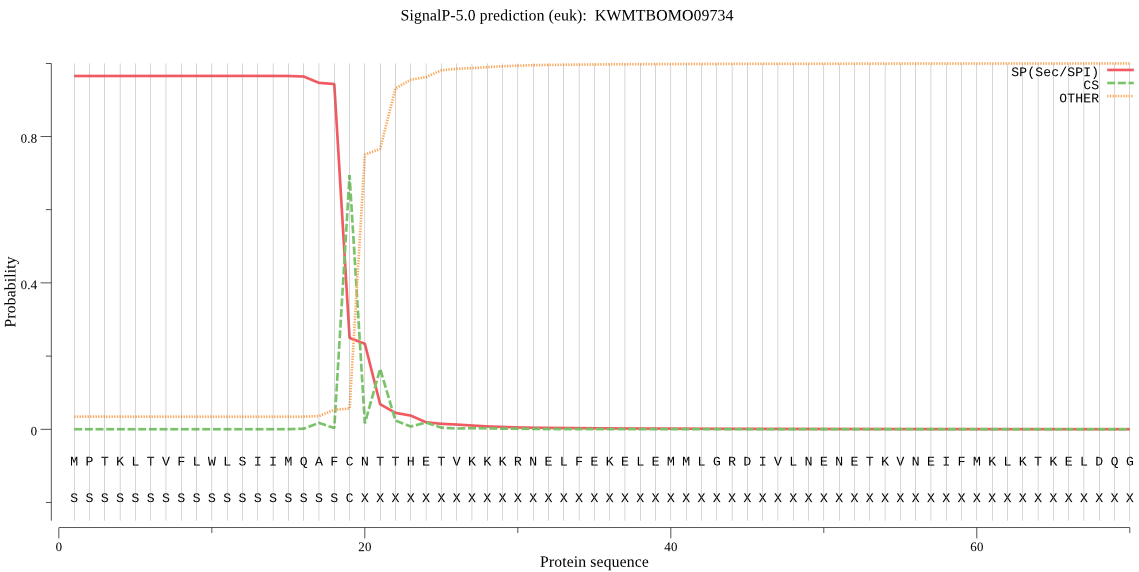
Length:
502
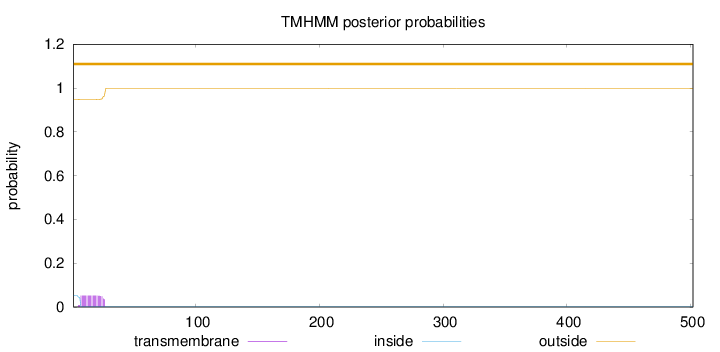
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.02211
Exp number, first 60 AAs:
1.02058
Total prob of N-in:
0.05221
outside
1 - 502
Population Genetic Test Statistics
Pi
279.200013
Theta
214.941656
Tajima's D
1.03855
CLR
0
CSRT
0.667716614169292
Interpretation
Uncertain
