Pre Gene Modal
BGIBMGA012893
Annotation
trafficking_protein_particle_complex_subunit_3_[Bombyx_mori]
Full name
Trafficking protein particle complex subunit
Location in the cell
Cytoplasmic Reliability : 2.063 Nuclear Reliability : 1.374
Sequence
CDS
ATGTCTCGGCAAACTTCTAGATTGGACGCCAAGAAAGTTAATTCCGAGCTCCTAACTCTTACATATGGGGCTTTAGTGTCTCAAATGCTGAAAGAAACAGAAAACACTGAAGATGTAAACAAACATCTTGAGAGAATCGGATACAACATGGGCGTTAGACTGATAGAGGACTTCCTGGCCCGGACTACGTCTACTCGCTGCTTAGAAATGAGAGAGACTGCTGATAAAATACAGCAGGCCTTCAAACTATACCTTAGCATGCAGCCGACAGTAACAAGTTGGAGTAGTGCCGGAGATGAGTTTTCGCTTGTTTGGGACCATTGTCCACTCAGTGAATGGGTGGAGATGCCAAGTAATAACGGCCTGAAATATTGCGCTTTGATTCCCGGTGCAATCAGAGGTGCTCTACAGATGGTACAACTAGATGTACAATGCTGGTTTGTGCAGGATCAGTTGAAAGGTGATCCCGTGACAGAATTAAGAGTGAAATACTTGAAGAGATTAGAAGATGCCGTACCAGCCGGTGAAGATTAG
Protein
MSRQTSRLDAKKVNSELLTLTYGALVSQMLKETENTEDVNKHLERIGYNMGVRLIEDFLARTTSTRCLEMRETADKIQQAFKLYLSMQPTVTSWSSAGDEFSLVWDHCPLSEWVEMPSNNGLKYCALIPGAIRGALQMVQLDVQCWFVQDQLKGDPVTELRVKYLKRLEDAVPAGED
Summary
Description
May play a role in vesicular transport from endoplasmic reticulum to Golgi.
Subunit
Homodimer.
Similarity
Belongs to the TRAPP small subunits family. BET3 subfamily.
Uniprot
Q2F629
A0A2W1BUB7
A0A2A4JDZ2
S4PF87
A0A1E1WK54
A0A212FMG2
+ More
A0A2H1VAQ3 A0A437B217 A0A194Q616 A0A194R2X6 H9JTN1 A0A1A9XIL9 B4MMU4 B4HKQ7 B3NCG3 B4QN65 Q9VSY8 B4LBR9 B4L994 A0A1W4U922 A0A0K8TTF7 B4PEW7 W8B247 A0A1I8NH44 A0A0K8UQ06 A0A3B0J5C4 B5DR31 B4H1Y5 A0A034VLG5 B3MA83 A0A1I8P740 A0A1Q3G2Y4 A0A1W7R891 Q0IEU7 A0A1S4FI35 U5EM23 A0A240PJY1 A0A1Y9HEN7 A0A1Y9IWG0 A0A336M1G9 A0A182M545 A0A1Y9J0E9 A0A1Y9H346 A0A1Y9HAS2 A0A1Y9IRY0 A0A2C9GTR9 A0A240PKB8 A0A1Y9IMI4 A0A1Y9GLV7 F5HIY1 A0A2K6VB53 A0A2M3ZAE2 A0A2M4AW20 A0A2M4C137 A0A1A9VN08 A0A1L8DVS3 B4J1I9 A0A182SDP7 A0A240PLN9 A0A2H8TXX8 C4WXG1 A0A1B0FAP2 V5GBT0 A0A026VUJ8 A0A1A9WRA1 B0XJC5 A0A1V9XYF0 D6WJE6 V9I956 A0A2P8Z8X1 B0WUG6 A0A0C9RTJ4 E0VBE5 A0A088AAJ1 A0A1Y1KY87 K7IQ15 A0A182HAW6 R4FPN5 A0A224XLU8 A0A0V0GBH1 A0A069DNS4 A0A158NTD7 V9IBF4 A0A0L0CIN0 A0A154PE55 A0A1B6GSY3 A0A1B6JGC9 A0A1B6M0F9 E9H4K9 A0A1W4X137 A0A0M4EDM9 N6U0W4 A0A0N8E0Z9 A0A0P6H0X2 A0A023F9H1 A0A2A3E1U3 A0A1S3CUN5 A0A2M4AWL1 A0A0P4X3L0 A0A182FDE7
A0A2H1VAQ3 A0A437B217 A0A194Q616 A0A194R2X6 H9JTN1 A0A1A9XIL9 B4MMU4 B4HKQ7 B3NCG3 B4QN65 Q9VSY8 B4LBR9 B4L994 A0A1W4U922 A0A0K8TTF7 B4PEW7 W8B247 A0A1I8NH44 A0A0K8UQ06 A0A3B0J5C4 B5DR31 B4H1Y5 A0A034VLG5 B3MA83 A0A1I8P740 A0A1Q3G2Y4 A0A1W7R891 Q0IEU7 A0A1S4FI35 U5EM23 A0A240PJY1 A0A1Y9HEN7 A0A1Y9IWG0 A0A336M1G9 A0A182M545 A0A1Y9J0E9 A0A1Y9H346 A0A1Y9HAS2 A0A1Y9IRY0 A0A2C9GTR9 A0A240PKB8 A0A1Y9IMI4 A0A1Y9GLV7 F5HIY1 A0A2K6VB53 A0A2M3ZAE2 A0A2M4AW20 A0A2M4C137 A0A1A9VN08 A0A1L8DVS3 B4J1I9 A0A182SDP7 A0A240PLN9 A0A2H8TXX8 C4WXG1 A0A1B0FAP2 V5GBT0 A0A026VUJ8 A0A1A9WRA1 B0XJC5 A0A1V9XYF0 D6WJE6 V9I956 A0A2P8Z8X1 B0WUG6 A0A0C9RTJ4 E0VBE5 A0A088AAJ1 A0A1Y1KY87 K7IQ15 A0A182HAW6 R4FPN5 A0A224XLU8 A0A0V0GBH1 A0A069DNS4 A0A158NTD7 V9IBF4 A0A0L0CIN0 A0A154PE55 A0A1B6GSY3 A0A1B6JGC9 A0A1B6M0F9 E9H4K9 A0A1W4X137 A0A0M4EDM9 N6U0W4 A0A0N8E0Z9 A0A0P6H0X2 A0A023F9H1 A0A2A3E1U3 A0A1S3CUN5 A0A2M4AWL1 A0A0P4X3L0 A0A182FDE7
Pubmed
28756777
23622113
22118469
26354079
19121390
17994087
+ More
22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26369729 17550304 24495485 25315136 15632085 25348373 17510324 20966253 12364791 14747013 17210077 20920257 24508170 30249741 28327890 18362917 19820115 29403074 20566863 28004739 20075255 26483478 26334808 21347285 26108605 21292972 23537049 25474469
22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26369729 17550304 24495485 25315136 15632085 25348373 17510324 20966253 12364791 14747013 17210077 20920257 24508170 30249741 28327890 18362917 19820115 29403074 20566863 28004739 20075255 26483478 26334808 21347285 26108605 21292972 23537049 25474469
EMBL
DQ311243
ABD36188.1
KZ149898
PZC78638.1
NWSH01001926
PCG69650.1
+ More
GAIX01006715 JAA85845.1 GDQN01003746 JAT87308.1 AGBW02007651 OWR54921.1 ODYU01001533 SOQ37881.1 RSAL01000205 RVE44354.1 KQ459582 KPI98850.1 KQ460878 KPJ11595.1 BABH01003477 CH963847 EDW73500.1 CH480815 EDW40860.1 CH954178 EDV51193.2 CM000363 CM002912 EDX09848.1 KMY98649.1 AE014296 BT015980 AAF50270.3 AAV36865.1 CH940647 EDW68696.2 CH933816 EDW17269.2 GDAI01000383 JAI17220.1 CM000159 EDW93022.2 GAMC01015457 JAB91098.1 GDHF01023555 JAI28759.1 OUUW01000002 SPP77154.1 CH379070 EDY74027.2 CH479203 EDW30337.1 GAKP01016310 GAKP01016307 GAKP01016305 GAKP01016301 JAC42645.1 CH902618 EDV39097.2 GFDL01000883 JAV34162.1 GEHC01000310 JAV47335.1 CH477478 EAT40254.1 GANO01001181 JAB58690.1 UFQT01000097 SSX19808.1 AXCM01000608 AXCN02000388 APCN01000197 AAAB01008807 EGK96242.1 GGFM01004753 MBW25504.1 GGFK01011668 MBW44989.1 GGFJ01009871 MBW59012.1 GFDF01003565 JAV10519.1 CH916366 EDV95880.1 GFXV01007348 MBW19153.1 ABLF02012718 AK342621 BAH72581.1 CCAG010020395 GALX01006997 JAB61469.1 KK107847 QOIP01000005 EZA47473.1 RLU22554.1 DS233480 EDS30162.1 MNPL01002098 OQR78547.1 KQ971342 EFA03885.1 JR037183 AEY57628.1 PYGN01000143 PSN52939.1 DS232104 EDS34945.1 GBYB01012065 JAG81832.1 DS235028 EEB10701.1 GEZM01070645 GEZM01070644 GEZM01070643 GEZM01070642 GEZM01070641 GEZM01070640 JAV66359.1 JXUM01031571 KQ560926 KXJ80318.1 ACPB03000027 GAHY01000789 JAA76721.1 GFTR01002960 JAW13466.1 GECL01000870 JAP05254.1 GBGD01003171 JAC85718.1 ADTU01025847 JR037184 AEY57629.1 JRES01000341 KNC32101.1 KQ434870 KZC09558.1 GECZ01004316 JAS65453.1 GECU01009436 JAS98270.1 GEBQ01014866 GEBQ01010576 GEBQ01009715 JAT25111.1 JAT29401.1 JAT30262.1 GL732592 EFX73158.1 CP012525 ALC43734.1 APGK01043984 KB741019 KB631645 ENN75165.1 ERL84969.1 GDIQ01072476 JAN22261.1 GDIQ01104857 GDIQ01026130 LRGB01000146 JAN68607.1 KZS20582.1 GBBI01000682 JAC18030.1 KZ288444 PBC25667.1 GGFK01011866 MBW45187.1 GDIP01249802 JAI73599.1
GAIX01006715 JAA85845.1 GDQN01003746 JAT87308.1 AGBW02007651 OWR54921.1 ODYU01001533 SOQ37881.1 RSAL01000205 RVE44354.1 KQ459582 KPI98850.1 KQ460878 KPJ11595.1 BABH01003477 CH963847 EDW73500.1 CH480815 EDW40860.1 CH954178 EDV51193.2 CM000363 CM002912 EDX09848.1 KMY98649.1 AE014296 BT015980 AAF50270.3 AAV36865.1 CH940647 EDW68696.2 CH933816 EDW17269.2 GDAI01000383 JAI17220.1 CM000159 EDW93022.2 GAMC01015457 JAB91098.1 GDHF01023555 JAI28759.1 OUUW01000002 SPP77154.1 CH379070 EDY74027.2 CH479203 EDW30337.1 GAKP01016310 GAKP01016307 GAKP01016305 GAKP01016301 JAC42645.1 CH902618 EDV39097.2 GFDL01000883 JAV34162.1 GEHC01000310 JAV47335.1 CH477478 EAT40254.1 GANO01001181 JAB58690.1 UFQT01000097 SSX19808.1 AXCM01000608 AXCN02000388 APCN01000197 AAAB01008807 EGK96242.1 GGFM01004753 MBW25504.1 GGFK01011668 MBW44989.1 GGFJ01009871 MBW59012.1 GFDF01003565 JAV10519.1 CH916366 EDV95880.1 GFXV01007348 MBW19153.1 ABLF02012718 AK342621 BAH72581.1 CCAG010020395 GALX01006997 JAB61469.1 KK107847 QOIP01000005 EZA47473.1 RLU22554.1 DS233480 EDS30162.1 MNPL01002098 OQR78547.1 KQ971342 EFA03885.1 JR037183 AEY57628.1 PYGN01000143 PSN52939.1 DS232104 EDS34945.1 GBYB01012065 JAG81832.1 DS235028 EEB10701.1 GEZM01070645 GEZM01070644 GEZM01070643 GEZM01070642 GEZM01070641 GEZM01070640 JAV66359.1 JXUM01031571 KQ560926 KXJ80318.1 ACPB03000027 GAHY01000789 JAA76721.1 GFTR01002960 JAW13466.1 GECL01000870 JAP05254.1 GBGD01003171 JAC85718.1 ADTU01025847 JR037184 AEY57629.1 JRES01000341 KNC32101.1 KQ434870 KZC09558.1 GECZ01004316 JAS65453.1 GECU01009436 JAS98270.1 GEBQ01014866 GEBQ01010576 GEBQ01009715 JAT25111.1 JAT29401.1 JAT30262.1 GL732592 EFX73158.1 CP012525 ALC43734.1 APGK01043984 KB741019 KB631645 ENN75165.1 ERL84969.1 GDIQ01072476 JAN22261.1 GDIQ01104857 GDIQ01026130 LRGB01000146 JAN68607.1 KZS20582.1 GBBI01000682 JAC18030.1 KZ288444 PBC25667.1 GGFK01011866 MBW45187.1 GDIP01249802 JAI73599.1
Proteomes
UP000218220
UP000007151
UP000283053
UP000053268
UP000053240
UP000005204
+ More
UP000092443 UP000007798 UP000001292 UP000008711 UP000000304 UP000000803 UP000008792 UP000009192 UP000192221 UP000002282 UP000095301 UP000268350 UP000001819 UP000008744 UP000007801 UP000095300 UP000008820 UP000075880 UP000075900 UP000075920 UP000075883 UP000076407 UP000075884 UP000075886 UP000075903 UP000075882 UP000075885 UP000075902 UP000075840 UP000007062 UP000078200 UP000001070 UP000075901 UP000075881 UP000007819 UP000092444 UP000053097 UP000279307 UP000091820 UP000002320 UP000192247 UP000007266 UP000245037 UP000009046 UP000005203 UP000002358 UP000069940 UP000249989 UP000015103 UP000005205 UP000037069 UP000076502 UP000000305 UP000192223 UP000092553 UP000019118 UP000030742 UP000076858 UP000242457 UP000079169 UP000069272
UP000092443 UP000007798 UP000001292 UP000008711 UP000000304 UP000000803 UP000008792 UP000009192 UP000192221 UP000002282 UP000095301 UP000268350 UP000001819 UP000008744 UP000007801 UP000095300 UP000008820 UP000075880 UP000075900 UP000075920 UP000075883 UP000076407 UP000075884 UP000075886 UP000075903 UP000075882 UP000075885 UP000075902 UP000075840 UP000007062 UP000078200 UP000001070 UP000075901 UP000075881 UP000007819 UP000092444 UP000053097 UP000279307 UP000091820 UP000002320 UP000192247 UP000007266 UP000245037 UP000009046 UP000005203 UP000002358 UP000069940 UP000249989 UP000015103 UP000005205 UP000037069 UP000076502 UP000000305 UP000192223 UP000092553 UP000019118 UP000030742 UP000076858 UP000242457 UP000079169 UP000069272
Interpro
SUPFAM
SSF111126
SSF111126
ProteinModelPortal
Q2F629
A0A2W1BUB7
A0A2A4JDZ2
S4PF87
A0A1E1WK54
A0A212FMG2
+ More
A0A2H1VAQ3 A0A437B217 A0A194Q616 A0A194R2X6 H9JTN1 A0A1A9XIL9 B4MMU4 B4HKQ7 B3NCG3 B4QN65 Q9VSY8 B4LBR9 B4L994 A0A1W4U922 A0A0K8TTF7 B4PEW7 W8B247 A0A1I8NH44 A0A0K8UQ06 A0A3B0J5C4 B5DR31 B4H1Y5 A0A034VLG5 B3MA83 A0A1I8P740 A0A1Q3G2Y4 A0A1W7R891 Q0IEU7 A0A1S4FI35 U5EM23 A0A240PJY1 A0A1Y9HEN7 A0A1Y9IWG0 A0A336M1G9 A0A182M545 A0A1Y9J0E9 A0A1Y9H346 A0A1Y9HAS2 A0A1Y9IRY0 A0A2C9GTR9 A0A240PKB8 A0A1Y9IMI4 A0A1Y9GLV7 F5HIY1 A0A2K6VB53 A0A2M3ZAE2 A0A2M4AW20 A0A2M4C137 A0A1A9VN08 A0A1L8DVS3 B4J1I9 A0A182SDP7 A0A240PLN9 A0A2H8TXX8 C4WXG1 A0A1B0FAP2 V5GBT0 A0A026VUJ8 A0A1A9WRA1 B0XJC5 A0A1V9XYF0 D6WJE6 V9I956 A0A2P8Z8X1 B0WUG6 A0A0C9RTJ4 E0VBE5 A0A088AAJ1 A0A1Y1KY87 K7IQ15 A0A182HAW6 R4FPN5 A0A224XLU8 A0A0V0GBH1 A0A069DNS4 A0A158NTD7 V9IBF4 A0A0L0CIN0 A0A154PE55 A0A1B6GSY3 A0A1B6JGC9 A0A1B6M0F9 E9H4K9 A0A1W4X137 A0A0M4EDM9 N6U0W4 A0A0N8E0Z9 A0A0P6H0X2 A0A023F9H1 A0A2A3E1U3 A0A1S3CUN5 A0A2M4AWL1 A0A0P4X3L0 A0A182FDE7
A0A2H1VAQ3 A0A437B217 A0A194Q616 A0A194R2X6 H9JTN1 A0A1A9XIL9 B4MMU4 B4HKQ7 B3NCG3 B4QN65 Q9VSY8 B4LBR9 B4L994 A0A1W4U922 A0A0K8TTF7 B4PEW7 W8B247 A0A1I8NH44 A0A0K8UQ06 A0A3B0J5C4 B5DR31 B4H1Y5 A0A034VLG5 B3MA83 A0A1I8P740 A0A1Q3G2Y4 A0A1W7R891 Q0IEU7 A0A1S4FI35 U5EM23 A0A240PJY1 A0A1Y9HEN7 A0A1Y9IWG0 A0A336M1G9 A0A182M545 A0A1Y9J0E9 A0A1Y9H346 A0A1Y9HAS2 A0A1Y9IRY0 A0A2C9GTR9 A0A240PKB8 A0A1Y9IMI4 A0A1Y9GLV7 F5HIY1 A0A2K6VB53 A0A2M3ZAE2 A0A2M4AW20 A0A2M4C137 A0A1A9VN08 A0A1L8DVS3 B4J1I9 A0A182SDP7 A0A240PLN9 A0A2H8TXX8 C4WXG1 A0A1B0FAP2 V5GBT0 A0A026VUJ8 A0A1A9WRA1 B0XJC5 A0A1V9XYF0 D6WJE6 V9I956 A0A2P8Z8X1 B0WUG6 A0A0C9RTJ4 E0VBE5 A0A088AAJ1 A0A1Y1KY87 K7IQ15 A0A182HAW6 R4FPN5 A0A224XLU8 A0A0V0GBH1 A0A069DNS4 A0A158NTD7 V9IBF4 A0A0L0CIN0 A0A154PE55 A0A1B6GSY3 A0A1B6JGC9 A0A1B6M0F9 E9H4K9 A0A1W4X137 A0A0M4EDM9 N6U0W4 A0A0N8E0Z9 A0A0P6H0X2 A0A023F9H1 A0A2A3E1U3 A0A1S3CUN5 A0A2M4AWL1 A0A0P4X3L0 A0A182FDE7
PDB
2J3W
E-value=1.01087e-55,
Score=544
Ontologies
GO
PANTHER
Topology
Subcellular location
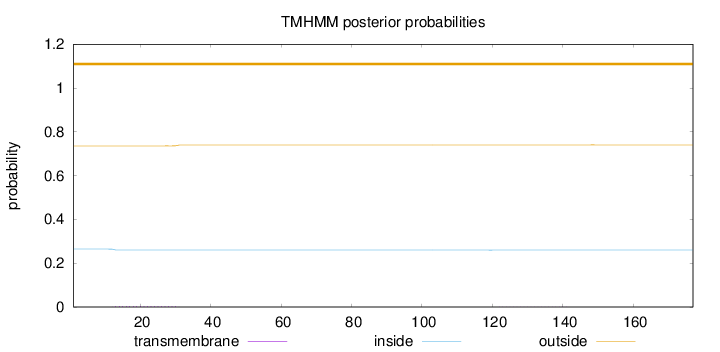
Length:
177
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0841099999999999
Exp number, first 60 AAs:
0.07612
Total prob of N-in:
0.26436
outside
1 - 177
Population Genetic Test Statistics
Pi
370.118973
Theta
218.308211
Tajima's D
2.158406
CLR
0
CSRT
0.904904754762262
Interpretation
Uncertain
