Gene
KWMTBOMO09726
Pre Gene Modal
BGIBMGA012892
Annotation
PREDICTED:_luciferin_4-monooxygenase-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.904
Sequence
CDS
ATGTGCTTGCCCACTTGTCCTGGTGAAACTGGAAAGGCCTGCGGGCCATCAATATCACCGCTTGAAATAGAGAAGGTCATTCGCCAACATCCAGGTGTGTTGGACGTCATAGTCACCAGCGTCACGGACGTCCAGCGCGAGGAGCTGCCGTGCGCTTGTGTCGTGCTTAAGGATGGTTGTCGCGTCACCGAACAGGAAATCAAGGACTTGGTTAAAGATTCGCTTTCGGATCCAAAACAATTGAGAGGCGGAGTTATTTTCCTAAAAGAAATGCCGACGACTCCTCAACTTAAAATTGACAGAAGGAAAATAAAGGAATTAGTGATTAACACTGTGAGGGAAGCATAA
Protein
MCLPTCPGETGKACGPSISPLEIEKVIRQHPGVLDVIVTSVTDVQREELPCACVVLKDGCRVTEQEIKDLVKDSLSDPKQLRGGVIFLKEMPTTPQLKIDRRKIKELVINTVREA
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
A0A0N1IQK8
A0A1B3TNX4
A0A1B3TP04
A0A194R6T5
A0A1B3TNY4
A0A194PZY0
+ More
A0A1B3TNZ3 A0A2W1BFJ2 A0A1B3TP14 A0A2A4JFU1 A0A2A4JDK9 A0A0L7LS68 A0A2W1BLY3 A0A2H1V3V7 A0A2A4JCF3 A0A2W1BJK0 A0A1B3TP07 A0A2A4JE16 A0A2A4J5M1 A0A1B3TP25 A0A2W1C0Z5 A0A2W1BJT9 A0A1B3TNY0 A0A2H1WV86 A0A2H1W204 A0A3S2N8I8 A0A2H1VFT6 A0A1B3TP10 A0A194Q640 A0A2A4JGT9 A0A1B3TNZ6 A0A1B3TNZ0 A0A2H1V0G3 H9JTS9 A0A1B3TNZ5 A0A2A4JKH2 A0A194Q0P0 A0A1B3TNX3 A0A212FBS4 A0A2A4J4G8 A0A2H1VAR1 A0A2H1VV91 A0A2H1V5T4 A0A2W1BGF2 A0A1B3TNZ2 A0A1B3TNX9 A0A1B3TP12 A0A1B3TNW9 A0A1B3TP22 A0A194PZW6 A0A194R232 A0A1B3TP24 A0A1B3TNX2 A0A212FN15 A0A0N1PKC2 A0A1B3TNZ4 A0A194R1K6 A0A2H1V5V8 A0A2H1VH10 A0A1B3TP11 A0A1B3TP01 A0A2H1WMC6 A0A2W1B861 A0A194Q1K5 A0A2A4JW63 A0A2H1VW29 A0A1B3TNY1 A0A1B0D5Z3 A0A1B3TNX0 A0A1B3TP03 A0A3S2NR21 A0A2A4JUE1 A0A1B3TP16 A0A212FBQ7 A0A194QZM0 A0A1L8DZ07 A0A1L8DY75 A0A1B3TNX7 H1AD93 A0A0L7L495 A0A2W1BC60 A0A3S2PGQ8 A0A418LUL6 A0A194PUZ5 A0A2H1VGY5 A0A2H1VNT1 A0A2T6WK65 A0A1B0DC38 A0A146MH35 A0A1L8DZ02 G9M6I9 A0A428D0L8 A0A2S3QZT6 A0A1L8DZ42 A0A1Q2T850 A0A1Q2U2F7 A0A194PT23 A0A1L8DZ97 A0A1B0GIH2
A0A1B3TNZ3 A0A2W1BFJ2 A0A1B3TP14 A0A2A4JFU1 A0A2A4JDK9 A0A0L7LS68 A0A2W1BLY3 A0A2H1V3V7 A0A2A4JCF3 A0A2W1BJK0 A0A1B3TP07 A0A2A4JE16 A0A2A4J5M1 A0A1B3TP25 A0A2W1C0Z5 A0A2W1BJT9 A0A1B3TNY0 A0A2H1WV86 A0A2H1W204 A0A3S2N8I8 A0A2H1VFT6 A0A1B3TP10 A0A194Q640 A0A2A4JGT9 A0A1B3TNZ6 A0A1B3TNZ0 A0A2H1V0G3 H9JTS9 A0A1B3TNZ5 A0A2A4JKH2 A0A194Q0P0 A0A1B3TNX3 A0A212FBS4 A0A2A4J4G8 A0A2H1VAR1 A0A2H1VV91 A0A2H1V5T4 A0A2W1BGF2 A0A1B3TNZ2 A0A1B3TNX9 A0A1B3TP12 A0A1B3TNW9 A0A1B3TP22 A0A194PZW6 A0A194R232 A0A1B3TP24 A0A1B3TNX2 A0A212FN15 A0A0N1PKC2 A0A1B3TNZ4 A0A194R1K6 A0A2H1V5V8 A0A2H1VH10 A0A1B3TP11 A0A1B3TP01 A0A2H1WMC6 A0A2W1B861 A0A194Q1K5 A0A2A4JW63 A0A2H1VW29 A0A1B3TNY1 A0A1B0D5Z3 A0A1B3TNX0 A0A1B3TP03 A0A3S2NR21 A0A2A4JUE1 A0A1B3TP16 A0A212FBQ7 A0A194QZM0 A0A1L8DZ07 A0A1L8DY75 A0A1B3TNX7 H1AD93 A0A0L7L495 A0A2W1BC60 A0A3S2PGQ8 A0A418LUL6 A0A194PUZ5 A0A2H1VGY5 A0A2H1VNT1 A0A2T6WK65 A0A1B0DC38 A0A146MH35 A0A1L8DZ02 G9M6I9 A0A428D0L8 A0A2S3QZT6 A0A1L8DZ42 A0A1Q2T850 A0A1Q2U2F7 A0A194PT23 A0A1L8DZ97 A0A1B0GIH2
EMBL
LADJ01029118
KPJ21198.1
KU925719
AOG75344.1
KU925757
AOG75382.1
+ More
KQ460878 KPJ11571.1 KU925729 AOG75354.1 KQ459582 KPI98876.1 KU925742 AOG75367.1 KZ150081 PZC73879.1 KU925772 AOG75397.1 NWSH01001632 PCG70636.1 NWSH01001893 PCG69778.1 JTDY01000201 KOB78320.1 PZC73876.1 ODYU01000351 SOQ34964.1 PCG69777.1 PZC73875.1 KU925761 AOG75386.1 NWSH01001926 PCG69652.1 NWSH01003054 PCG67036.1 KU925774 AOG75399.1 KZ149898 PZC78636.1 KZ150128 PZC73096.1 KU925732 AOG75357.1 ODYU01011279 SOQ56917.1 ODYU01005841 SOQ47125.1 RSAL01000205 RVE44356.1 ODYU01002332 SOQ39697.1 KU925762 AOG75387.1 KPI98875.1 PCG70633.1 KU925747 AOG75372.1 KU925745 AOG75370.1 ODYU01000111 SOQ34327.1 BABH01003491 BABH01003492 KU925746 AOG75371.1 NWSH01001271 PCG71900.1 KPI98873.1 KU925722 AOG75347.1 AGBW02009293 OWR51173.1 PCG67037.1 ODYU01001291 SOQ37304.1 ODYU01004651 SOQ44755.1 ODYU01000839 SOQ36210.1 PZC73878.1 KU925733 AOG75358.1 KU925730 AOG75355.1 KU925758 AOG75383.1 KU925721 AOG75346.1 KU925773 AOG75398.1 KPI98872.1 KPJ11574.1 KU925775 AOG75400.1 KU925720 AOG75345.1 AGBW02007648 OWR55128.1 KPJ21196.1 KU925743 AOG75368.1 KPJ11572.1 ODYU01000840 SOQ36211.1 ODYU01002515 SOQ40108.1 KU925759 AOG75384.1 KU925744 AOG75369.1 ODYU01009613 SOQ54167.1 KZ150230 PZC72009.1 KPI98874.1 NWSH01000564 PCG75622.1 ODYU01004634 SOQ44712.1 KU925740 AOG75365.1 AJVK01003542 AJVK01003543 AJVK01003544 AJVK01003545 KU925717 AOG75342.1 KU925755 AOG75380.1 RSAL01000342 RVE42388.1 PCG75621.1 KU925770 AOG75395.1 OWR51172.1 KQ460890 KPJ10988.1 GFDF01002418 JAV11666.1 GFDF01002674 JAV11410.1 KU925727 AOG75352.1 AB644225 BAL46509.1 JTDY01003125 KOB70109.1 PZC72011.1 RSAL01000040 RVE50945.1 QXHL01000184 RIV16326.1 KQ459593 KPI96564.1 SOQ40110.1 ODYU01003402 SOQ42112.1 PYJI01000254 PUE89139.1 AJVK01004974 GDHC01000649 JAQ17980.1 GFDF01002383 JAV11701.1 AB604790 BAL40875.1 RJNV01000052 RSI85664.1 PDGH01000119 POB45175.1 GFDF01002351 JAV11733.1 LC198321 BAW87787.1 LC215694 BAW98153.1 KPI96566.1 GFDF01002337 JAV11747.1 AJWK01014116
KQ460878 KPJ11571.1 KU925729 AOG75354.1 KQ459582 KPI98876.1 KU925742 AOG75367.1 KZ150081 PZC73879.1 KU925772 AOG75397.1 NWSH01001632 PCG70636.1 NWSH01001893 PCG69778.1 JTDY01000201 KOB78320.1 PZC73876.1 ODYU01000351 SOQ34964.1 PCG69777.1 PZC73875.1 KU925761 AOG75386.1 NWSH01001926 PCG69652.1 NWSH01003054 PCG67036.1 KU925774 AOG75399.1 KZ149898 PZC78636.1 KZ150128 PZC73096.1 KU925732 AOG75357.1 ODYU01011279 SOQ56917.1 ODYU01005841 SOQ47125.1 RSAL01000205 RVE44356.1 ODYU01002332 SOQ39697.1 KU925762 AOG75387.1 KPI98875.1 PCG70633.1 KU925747 AOG75372.1 KU925745 AOG75370.1 ODYU01000111 SOQ34327.1 BABH01003491 BABH01003492 KU925746 AOG75371.1 NWSH01001271 PCG71900.1 KPI98873.1 KU925722 AOG75347.1 AGBW02009293 OWR51173.1 PCG67037.1 ODYU01001291 SOQ37304.1 ODYU01004651 SOQ44755.1 ODYU01000839 SOQ36210.1 PZC73878.1 KU925733 AOG75358.1 KU925730 AOG75355.1 KU925758 AOG75383.1 KU925721 AOG75346.1 KU925773 AOG75398.1 KPI98872.1 KPJ11574.1 KU925775 AOG75400.1 KU925720 AOG75345.1 AGBW02007648 OWR55128.1 KPJ21196.1 KU925743 AOG75368.1 KPJ11572.1 ODYU01000840 SOQ36211.1 ODYU01002515 SOQ40108.1 KU925759 AOG75384.1 KU925744 AOG75369.1 ODYU01009613 SOQ54167.1 KZ150230 PZC72009.1 KPI98874.1 NWSH01000564 PCG75622.1 ODYU01004634 SOQ44712.1 KU925740 AOG75365.1 AJVK01003542 AJVK01003543 AJVK01003544 AJVK01003545 KU925717 AOG75342.1 KU925755 AOG75380.1 RSAL01000342 RVE42388.1 PCG75621.1 KU925770 AOG75395.1 OWR51172.1 KQ460890 KPJ10988.1 GFDF01002418 JAV11666.1 GFDF01002674 JAV11410.1 KU925727 AOG75352.1 AB644225 BAL46509.1 JTDY01003125 KOB70109.1 PZC72011.1 RSAL01000040 RVE50945.1 QXHL01000184 RIV16326.1 KQ459593 KPI96564.1 SOQ40110.1 ODYU01003402 SOQ42112.1 PYJI01000254 PUE89139.1 AJVK01004974 GDHC01000649 JAQ17980.1 GFDF01002383 JAV11701.1 AB604790 BAL40875.1 RJNV01000052 RSI85664.1 PDGH01000119 POB45175.1 GFDF01002351 JAV11733.1 LC198321 BAW87787.1 LC215694 BAW98153.1 KPI96566.1 GFDF01002337 JAV11747.1 AJWK01014116
Proteomes
PRIDE
Interpro
IPR016696
TRAPP-I_su5
+ More
IPR007194 TRAPP_component
IPR000873 AMP-dep_Synth/Lig
IPR025110 AMP-bd_C
IPR024096 NO_sig/Golgi_transp_ligand-bd
IPR042099 AMP-dep_Synthh-like_sf
IPR020845 AMP-binding_CS
IPR027380 Luciferase-like_N_sf
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR033116 TRYPSIN_SER
IPR007194 TRAPP_component
IPR000873 AMP-dep_Synth/Lig
IPR025110 AMP-bd_C
IPR024096 NO_sig/Golgi_transp_ligand-bd
IPR042099 AMP-dep_Synthh-like_sf
IPR020845 AMP-binding_CS
IPR027380 Luciferase-like_N_sf
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR033116 TRYPSIN_SER
Gene 3D
ProteinModelPortal
A0A0N1IQK8
A0A1B3TNX4
A0A1B3TP04
A0A194R6T5
A0A1B3TNY4
A0A194PZY0
+ More
A0A1B3TNZ3 A0A2W1BFJ2 A0A1B3TP14 A0A2A4JFU1 A0A2A4JDK9 A0A0L7LS68 A0A2W1BLY3 A0A2H1V3V7 A0A2A4JCF3 A0A2W1BJK0 A0A1B3TP07 A0A2A4JE16 A0A2A4J5M1 A0A1B3TP25 A0A2W1C0Z5 A0A2W1BJT9 A0A1B3TNY0 A0A2H1WV86 A0A2H1W204 A0A3S2N8I8 A0A2H1VFT6 A0A1B3TP10 A0A194Q640 A0A2A4JGT9 A0A1B3TNZ6 A0A1B3TNZ0 A0A2H1V0G3 H9JTS9 A0A1B3TNZ5 A0A2A4JKH2 A0A194Q0P0 A0A1B3TNX3 A0A212FBS4 A0A2A4J4G8 A0A2H1VAR1 A0A2H1VV91 A0A2H1V5T4 A0A2W1BGF2 A0A1B3TNZ2 A0A1B3TNX9 A0A1B3TP12 A0A1B3TNW9 A0A1B3TP22 A0A194PZW6 A0A194R232 A0A1B3TP24 A0A1B3TNX2 A0A212FN15 A0A0N1PKC2 A0A1B3TNZ4 A0A194R1K6 A0A2H1V5V8 A0A2H1VH10 A0A1B3TP11 A0A1B3TP01 A0A2H1WMC6 A0A2W1B861 A0A194Q1K5 A0A2A4JW63 A0A2H1VW29 A0A1B3TNY1 A0A1B0D5Z3 A0A1B3TNX0 A0A1B3TP03 A0A3S2NR21 A0A2A4JUE1 A0A1B3TP16 A0A212FBQ7 A0A194QZM0 A0A1L8DZ07 A0A1L8DY75 A0A1B3TNX7 H1AD93 A0A0L7L495 A0A2W1BC60 A0A3S2PGQ8 A0A418LUL6 A0A194PUZ5 A0A2H1VGY5 A0A2H1VNT1 A0A2T6WK65 A0A1B0DC38 A0A146MH35 A0A1L8DZ02 G9M6I9 A0A428D0L8 A0A2S3QZT6 A0A1L8DZ42 A0A1Q2T850 A0A1Q2U2F7 A0A194PT23 A0A1L8DZ97 A0A1B0GIH2
A0A1B3TNZ3 A0A2W1BFJ2 A0A1B3TP14 A0A2A4JFU1 A0A2A4JDK9 A0A0L7LS68 A0A2W1BLY3 A0A2H1V3V7 A0A2A4JCF3 A0A2W1BJK0 A0A1B3TP07 A0A2A4JE16 A0A2A4J5M1 A0A1B3TP25 A0A2W1C0Z5 A0A2W1BJT9 A0A1B3TNY0 A0A2H1WV86 A0A2H1W204 A0A3S2N8I8 A0A2H1VFT6 A0A1B3TP10 A0A194Q640 A0A2A4JGT9 A0A1B3TNZ6 A0A1B3TNZ0 A0A2H1V0G3 H9JTS9 A0A1B3TNZ5 A0A2A4JKH2 A0A194Q0P0 A0A1B3TNX3 A0A212FBS4 A0A2A4J4G8 A0A2H1VAR1 A0A2H1VV91 A0A2H1V5T4 A0A2W1BGF2 A0A1B3TNZ2 A0A1B3TNX9 A0A1B3TP12 A0A1B3TNW9 A0A1B3TP22 A0A194PZW6 A0A194R232 A0A1B3TP24 A0A1B3TNX2 A0A212FN15 A0A0N1PKC2 A0A1B3TNZ4 A0A194R1K6 A0A2H1V5V8 A0A2H1VH10 A0A1B3TP11 A0A1B3TP01 A0A2H1WMC6 A0A2W1B861 A0A194Q1K5 A0A2A4JW63 A0A2H1VW29 A0A1B3TNY1 A0A1B0D5Z3 A0A1B3TNX0 A0A1B3TP03 A0A3S2NR21 A0A2A4JUE1 A0A1B3TP16 A0A212FBQ7 A0A194QZM0 A0A1L8DZ07 A0A1L8DY75 A0A1B3TNX7 H1AD93 A0A0L7L495 A0A2W1BC60 A0A3S2PGQ8 A0A418LUL6 A0A194PUZ5 A0A2H1VGY5 A0A2H1VNT1 A0A2T6WK65 A0A1B0DC38 A0A146MH35 A0A1L8DZ02 G9M6I9 A0A428D0L8 A0A2S3QZT6 A0A1L8DZ42 A0A1Q2T850 A0A1Q2U2F7 A0A194PT23 A0A1L8DZ97 A0A1B0GIH2
PDB
4G37
E-value=1.157e-16,
Score=205
Ontologies
PANTHER
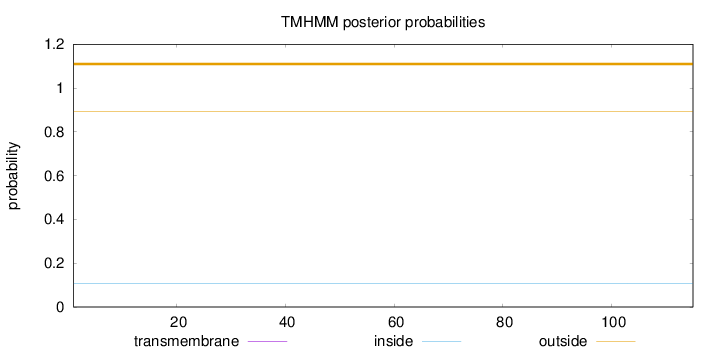
Topology
Length:
115
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.10826
outside
1 - 115
Population Genetic Test Statistics
Pi
120.45274
Theta
126.332354
Tajima's D
-0.532927
CLR
6.785916
CSRT
0.230988450577471
Interpretation
Uncertain
