Gene
KWMTBOMO09725
Pre Gene Modal
BGIBMGA012892
Annotation
PREDICTED:_luciferin_4-monooxygenase-like_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.554
Sequence
CDS
ATGGAGATGACAGACAATGCACTTGCACAAATTTTCGCTTCAAACCAACCAAAAGCTATATTTTGTCAGAACAGAAATCTTGCTAACGTAAAAAATGCACTGGAGTTTTATGAGCATGATATAAAATTACTTACCTTCGATGATAATGATCAAAAGGACGTTTTAACTTTTTCTGAATTGATGACTGGTTTTGGTAGTGATGCATCGATTGGAAGCTTCAAGCCAACAGATTTTGATCCCGAAGAAACAATTGCTTTGATAACTGAAACAAGTGGAACTACTGGAGCGCCAAAGGGCGTTGCTCTTACGCACAAAAATTTAGTCATATCAATTACTTTTAATCGGTAA
Protein
MEMTDNALAQIFASNQPKAIFCQNRNLANVKNALEFYEHDIKLLTFDDNDQKDVLTFSELMTGFGSDASIGSFKPTDFDPEETIALITETSGTTGAPKGVALTHKNLVISITFNR
Summary
Uniprot
H9JTN0
H9JTM9
A0A1B3TNZ4
A0A1B3TP22
A0A1B3TP12
A0A194PZY0
+ More
A0A1B3TNX9 A0A2H1VVH2 A0A1B3TNX2 A0A194R6T5 A0A212FN15 A0A2A4JDK9 A0A2H1WV86 A0A1B3TP07 A0A2H1V5V8 A0A2A4JEW6 A0A2H1VAR1 A0A2H1VV91 A0A1B3TNZ0 A0A2W1BGF2 A0A2A4JE16 A0A2H1V5T4 A0A2A4JKH2 A0A2W1BJT9 A0A2W1BJK0 A0A2A4JFU1 A0A2A4J4G8 A0A1B3TP11 A0A1B3TNX4 A0A1B3TNX3 A0A2A4JCF3 A0A2W1C0Z5 A0A2W1BLY3 A0A2H1W204 A0A1B3TP24 A0A1B3TP14 A0A0L7LS68 A0A2H1V297 A0A1B3TNZ2 A0A1B3TNY4 A0A2A4JGC8 A0A194R232 A0A2A4J5M1 A0A1B3TNZ3 A0A194Q1K5 A0A3S2N8I8 A0A1B3TP04 A0A1B3TP10 A0A1B3TNY0 A0A1B3TP09 A0A1B3TP25 A0A2H1VFT6 A0A1B3TP01 A0A194R1K6 A0A2W1BME0 A0A194PZW6 A0A1B3TNZ6 A0A194Q640 A0A1B3TNW9 H9JTS9 A0A1B3TNZ5 A0A212FBS4 A0A0L7LJN8 A0A194Q0P0 Q05KC0 A0A0C6E621 A0A0C6DRW5 D2KV87 A0A1Y1LAA7 Q2ACC8 Q1ET68 A0A1H8DB51 D2KV86 E9JGN2 A0A1J6HUA1 D2DHG8 Q9LKM8 E9REL2 E3WI51 A0A1W4WUS0 A0A1W4X5G4 A0A1W4WHZ8 D2DHG9 A0A1W4X5M1 Q718F0 A0A1Y1N7D9 D6WTG3 Q9U4U8 D6WTM3 Q718E3 Q718F1 A0A0T6B4L2 A0A0C6E0Y4 D2KV85 Q9LL50 M1B5G9 Q718D1
A0A1B3TNX9 A0A2H1VVH2 A0A1B3TNX2 A0A194R6T5 A0A212FN15 A0A2A4JDK9 A0A2H1WV86 A0A1B3TP07 A0A2H1V5V8 A0A2A4JEW6 A0A2H1VAR1 A0A2H1VV91 A0A1B3TNZ0 A0A2W1BGF2 A0A2A4JE16 A0A2H1V5T4 A0A2A4JKH2 A0A2W1BJT9 A0A2W1BJK0 A0A2A4JFU1 A0A2A4J4G8 A0A1B3TP11 A0A1B3TNX4 A0A1B3TNX3 A0A2A4JCF3 A0A2W1C0Z5 A0A2W1BLY3 A0A2H1W204 A0A1B3TP24 A0A1B3TP14 A0A0L7LS68 A0A2H1V297 A0A1B3TNZ2 A0A1B3TNY4 A0A2A4JGC8 A0A194R232 A0A2A4J5M1 A0A1B3TNZ3 A0A194Q1K5 A0A3S2N8I8 A0A1B3TP04 A0A1B3TP10 A0A1B3TNY0 A0A1B3TP09 A0A1B3TP25 A0A2H1VFT6 A0A1B3TP01 A0A194R1K6 A0A2W1BME0 A0A194PZW6 A0A1B3TNZ6 A0A194Q640 A0A1B3TNW9 H9JTS9 A0A1B3TNZ5 A0A212FBS4 A0A0L7LJN8 A0A194Q0P0 Q05KC0 A0A0C6E621 A0A0C6DRW5 D2KV87 A0A1Y1LAA7 Q2ACC8 Q1ET68 A0A1H8DB51 D2KV86 E9JGN2 A0A1J6HUA1 D2DHG8 Q9LKM8 E9REL2 E3WI51 A0A1W4WUS0 A0A1W4X5G4 A0A1W4WHZ8 D2DHG9 A0A1W4X5M1 Q718F0 A0A1Y1N7D9 D6WTG3 Q9U4U8 D6WTM3 Q718E3 Q718F1 A0A0T6B4L2 A0A0C6E0Y4 D2KV85 Q9LL50 M1B5G9 Q718D1
Pubmed
EMBL
BABH01003478
BABH01003479
BABH01003480
BABH01003481
BABH01003489
KU925743
+ More
AOG75368.1 KU925773 AOG75398.1 KU925758 AOG75383.1 KQ459582 KPI98876.1 KU925730 AOG75355.1 ODYU01004651 SOQ44756.1 KU925720 AOG75345.1 KQ460878 KPJ11571.1 AGBW02007648 OWR55128.1 NWSH01001893 PCG69778.1 ODYU01011279 SOQ56917.1 KU925761 AOG75386.1 ODYU01000840 SOQ36211.1 NWSH01001632 PCG70635.1 ODYU01001291 SOQ37304.1 SOQ44755.1 KU925745 AOG75370.1 KZ150081 PZC73878.1 NWSH01001926 PCG69652.1 ODYU01000839 SOQ36210.1 NWSH01001271 PCG71900.1 KZ150128 PZC73096.1 PZC73875.1 PCG70636.1 NWSH01003054 PCG67037.1 KU925759 AOG75384.1 KU925719 AOG75344.1 KU925722 AOG75347.1 PCG69777.1 KZ149898 PZC78636.1 PZC73876.1 ODYU01005841 SOQ47125.1 KU925775 AOG75400.1 KU925772 AOG75397.1 JTDY01000201 KOB78320.1 ODYU01000351 SOQ34963.1 KU925733 AOG75358.1 KU925729 AOG75354.1 PCG70634.1 KPJ11574.1 PCG67036.1 KU925742 AOG75367.1 KPI98874.1 RSAL01000205 RVE44356.1 KU925757 AOG75382.1 KU925762 AOG75387.1 KU925732 AOG75357.1 KU925760 AOG75385.1 KU925774 AOG75399.1 ODYU01002332 SOQ39697.1 KU925744 AOG75369.1 KPJ11572.1 PZC73880.1 KPI98872.1 KU925747 AOG75372.1 KPI98875.1 KU925721 AOG75346.1 BABH01003491 BABH01003492 KU925746 AOG75371.1 AGBW02009293 OWR51173.1 JTDY01000923 KOB75416.1 KPI98873.1 AB255748 BAF34360.1 AB251886 BAQ25863.1 AB251887 BAQ25864.1 AB479114 BAI66602.1 GEZM01063972 JAV68885.1 AB196456 BAE80729.1 AB098617 BAE95691.1 FOBW01000008 SEN04601.1 AB479113 BAI66601.1 GU117606 ADB25236.1 MJEQ01037194 OIS96445.1 FJ545728 ACT68596.1 AF270934 AAF78074.1 AB490793 BAJ83485.1 AB603804 BAJ41368.1 FJ545729 ACT68597.1 AF543368 AF543370 AF543371 AF543372 AF543373 AF543374 AF543375 AF543376 AAQ11690.1 GEZM01010792 JAV93832.1 KQ971352 EFA06717.1 AF139644 AAD34542.1 KQ971355 EFA05828.2 AF543377 AAQ11697.1 AF543369 AAQ11689.1 LJIG01009802 KRT82352.1 AB251885 BAQ25862.1 AB479112 BAI66600.1 AF239686 AAF91309.1 AF543388 AF543389 AF543390 AF543391 AF543394 AAQ11709.1
AOG75368.1 KU925773 AOG75398.1 KU925758 AOG75383.1 KQ459582 KPI98876.1 KU925730 AOG75355.1 ODYU01004651 SOQ44756.1 KU925720 AOG75345.1 KQ460878 KPJ11571.1 AGBW02007648 OWR55128.1 NWSH01001893 PCG69778.1 ODYU01011279 SOQ56917.1 KU925761 AOG75386.1 ODYU01000840 SOQ36211.1 NWSH01001632 PCG70635.1 ODYU01001291 SOQ37304.1 SOQ44755.1 KU925745 AOG75370.1 KZ150081 PZC73878.1 NWSH01001926 PCG69652.1 ODYU01000839 SOQ36210.1 NWSH01001271 PCG71900.1 KZ150128 PZC73096.1 PZC73875.1 PCG70636.1 NWSH01003054 PCG67037.1 KU925759 AOG75384.1 KU925719 AOG75344.1 KU925722 AOG75347.1 PCG69777.1 KZ149898 PZC78636.1 PZC73876.1 ODYU01005841 SOQ47125.1 KU925775 AOG75400.1 KU925772 AOG75397.1 JTDY01000201 KOB78320.1 ODYU01000351 SOQ34963.1 KU925733 AOG75358.1 KU925729 AOG75354.1 PCG70634.1 KPJ11574.1 PCG67036.1 KU925742 AOG75367.1 KPI98874.1 RSAL01000205 RVE44356.1 KU925757 AOG75382.1 KU925762 AOG75387.1 KU925732 AOG75357.1 KU925760 AOG75385.1 KU925774 AOG75399.1 ODYU01002332 SOQ39697.1 KU925744 AOG75369.1 KPJ11572.1 PZC73880.1 KPI98872.1 KU925747 AOG75372.1 KPI98875.1 KU925721 AOG75346.1 BABH01003491 BABH01003492 KU925746 AOG75371.1 AGBW02009293 OWR51173.1 JTDY01000923 KOB75416.1 KPI98873.1 AB255748 BAF34360.1 AB251886 BAQ25863.1 AB251887 BAQ25864.1 AB479114 BAI66602.1 GEZM01063972 JAV68885.1 AB196456 BAE80729.1 AB098617 BAE95691.1 FOBW01000008 SEN04601.1 AB479113 BAI66601.1 GU117606 ADB25236.1 MJEQ01037194 OIS96445.1 FJ545728 ACT68596.1 AF270934 AAF78074.1 AB490793 BAJ83485.1 AB603804 BAJ41368.1 FJ545729 ACT68597.1 AF543368 AF543370 AF543371 AF543372 AF543373 AF543374 AF543375 AF543376 AAQ11690.1 GEZM01010792 JAV93832.1 KQ971352 EFA06717.1 AF139644 AAD34542.1 KQ971355 EFA05828.2 AF543377 AAQ11697.1 AF543369 AAQ11689.1 LJIG01009802 KRT82352.1 AB251885 BAQ25862.1 AB479112 BAI66600.1 AF239686 AAF91309.1 AF543388 AF543389 AF543390 AF543391 AF543394 AAQ11709.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
H9JTN0
H9JTM9
A0A1B3TNZ4
A0A1B3TP22
A0A1B3TP12
A0A194PZY0
+ More
A0A1B3TNX9 A0A2H1VVH2 A0A1B3TNX2 A0A194R6T5 A0A212FN15 A0A2A4JDK9 A0A2H1WV86 A0A1B3TP07 A0A2H1V5V8 A0A2A4JEW6 A0A2H1VAR1 A0A2H1VV91 A0A1B3TNZ0 A0A2W1BGF2 A0A2A4JE16 A0A2H1V5T4 A0A2A4JKH2 A0A2W1BJT9 A0A2W1BJK0 A0A2A4JFU1 A0A2A4J4G8 A0A1B3TP11 A0A1B3TNX4 A0A1B3TNX3 A0A2A4JCF3 A0A2W1C0Z5 A0A2W1BLY3 A0A2H1W204 A0A1B3TP24 A0A1B3TP14 A0A0L7LS68 A0A2H1V297 A0A1B3TNZ2 A0A1B3TNY4 A0A2A4JGC8 A0A194R232 A0A2A4J5M1 A0A1B3TNZ3 A0A194Q1K5 A0A3S2N8I8 A0A1B3TP04 A0A1B3TP10 A0A1B3TNY0 A0A1B3TP09 A0A1B3TP25 A0A2H1VFT6 A0A1B3TP01 A0A194R1K6 A0A2W1BME0 A0A194PZW6 A0A1B3TNZ6 A0A194Q640 A0A1B3TNW9 H9JTS9 A0A1B3TNZ5 A0A212FBS4 A0A0L7LJN8 A0A194Q0P0 Q05KC0 A0A0C6E621 A0A0C6DRW5 D2KV87 A0A1Y1LAA7 Q2ACC8 Q1ET68 A0A1H8DB51 D2KV86 E9JGN2 A0A1J6HUA1 D2DHG8 Q9LKM8 E9REL2 E3WI51 A0A1W4WUS0 A0A1W4X5G4 A0A1W4WHZ8 D2DHG9 A0A1W4X5M1 Q718F0 A0A1Y1N7D9 D6WTG3 Q9U4U8 D6WTM3 Q718E3 Q718F1 A0A0T6B4L2 A0A0C6E0Y4 D2KV85 Q9LL50 M1B5G9 Q718D1
A0A1B3TNX9 A0A2H1VVH2 A0A1B3TNX2 A0A194R6T5 A0A212FN15 A0A2A4JDK9 A0A2H1WV86 A0A1B3TP07 A0A2H1V5V8 A0A2A4JEW6 A0A2H1VAR1 A0A2H1VV91 A0A1B3TNZ0 A0A2W1BGF2 A0A2A4JE16 A0A2H1V5T4 A0A2A4JKH2 A0A2W1BJT9 A0A2W1BJK0 A0A2A4JFU1 A0A2A4J4G8 A0A1B3TP11 A0A1B3TNX4 A0A1B3TNX3 A0A2A4JCF3 A0A2W1C0Z5 A0A2W1BLY3 A0A2H1W204 A0A1B3TP24 A0A1B3TP14 A0A0L7LS68 A0A2H1V297 A0A1B3TNZ2 A0A1B3TNY4 A0A2A4JGC8 A0A194R232 A0A2A4J5M1 A0A1B3TNZ3 A0A194Q1K5 A0A3S2N8I8 A0A1B3TP04 A0A1B3TP10 A0A1B3TNY0 A0A1B3TP09 A0A1B3TP25 A0A2H1VFT6 A0A1B3TP01 A0A194R1K6 A0A2W1BME0 A0A194PZW6 A0A1B3TNZ6 A0A194Q640 A0A1B3TNW9 H9JTS9 A0A1B3TNZ5 A0A212FBS4 A0A0L7LJN8 A0A194Q0P0 Q05KC0 A0A0C6E621 A0A0C6DRW5 D2KV87 A0A1Y1LAA7 Q2ACC8 Q1ET68 A0A1H8DB51 D2KV86 E9JGN2 A0A1J6HUA1 D2DHG8 Q9LKM8 E9REL2 E3WI51 A0A1W4WUS0 A0A1W4X5G4 A0A1W4WHZ8 D2DHG9 A0A1W4X5M1 Q718F0 A0A1Y1N7D9 D6WTG3 Q9U4U8 D6WTM3 Q718E3 Q718F1 A0A0T6B4L2 A0A0C6E0Y4 D2KV85 Q9LL50 M1B5G9 Q718D1
PDB
6AC3
E-value=2.54076e-08,
Score=133
Ontologies
GO
Topology
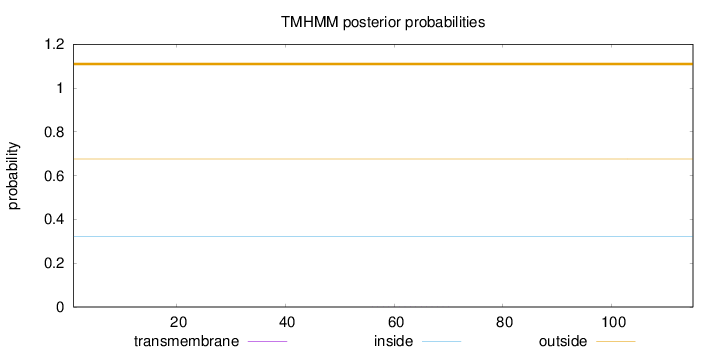
Length:
115
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00404
Exp number, first 60 AAs:
0.00081
Total prob of N-in:
0.32356
outside
1 - 115
Population Genetic Test Statistics
Pi
286.450719
Theta
204.737139
Tajima's D
1.078818
CLR
0.000048
CSRT
0.67941602919854
Interpretation
Uncertain
