Gene
KWMTBOMO09720 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012890
Annotation
PREDICTED:_fatty_acid-binding_protein_1-like_isoform_X2_[Bombyx_mori]
Full name
Fatty acid-binding protein 1
+ More
Fatty acid-binding protein 2
Fatty acid-binding protein 2
Location in the cell
Cytoplasmic Reliability : 3.174
Sequence
CDS
ATGTCTTTCTTGGGTAAAAAATACACATTAGATAGAGAAGAAAACTTCGATGGGTTCCTTAAGTTTATCGGTCTCCCCGAAGACCAGATCCAGAAACATTCGCAGTTCAAGCCTACAGCTGTATTGACCAAGGAAGGCAACAAATACAAGTCCATCACCGTCAACACCGACGGTCCAAAGGAGTCAGTTTTTGAGTCCGGTGTGCCTTTCGACGAAGTCGTCCCAGGTGGATTCAAGGTAAAAACCATGTACATAGTCGATGGCAACACCGTAACTCAGACTGTAGAGAATCCCAATGGCATAGCAACATTCAAAAGGGAATACAGTGGCAACGAACTAAAAGTTACTGTCACAGCCGATAAATGGGACGGCACTGCCTATCGATACTACAAAGCTTAA
Protein
MSFLGKKYTLDREENFDGFLKFIGLPEDQIQKHSQFKPTAVLTKEGNKYKSITVNTDGPKESVFESGVPFDEVVPGGFKVKTMYIVDGNTVTQTVENPNGIATFKREYSGNELKVTVTADKWDGTAYRYYKA
Summary
Subunit
Monomer.
Similarity
Belongs to the calycin superfamily. Fatty-acid binding protein (FABP) family.
Keywords
Cytoplasm
Lipid-binding
Transport
3D-structure
Acetylation
Feature
chain Fatty acid-binding protein 1
Uniprot
H9JTM8
Q2F5R6
H9JTT1
P31416
A0A2W1BFP9
A0A2A4IYF3
+ More
B6CMF8 A0A2H1VZX6 P31417 A0A2A4IXI1 A0A2H1VZX7 A0A2H1V0J1 B6CMF9 A0A2A4IZR3 A0A2A4IZ24 A0A2W1BCW9 A0A2W1BK37 F8URE4 A0A2W1BCW8 A0A2A4IY62 A0A194Q1K0 A0A2H1VZW5 A0A2H1V1V6 A0A212FN08 A0A194R6U0 A0A2A4IYX0 A0A212FN12 Q2PWA4 A0A2H1W0N8 A0A194Q0Y2 A0A0N1IFX3 A0A2W1BHC4 A0A0N1I994 A0A1E1WE07 A0A3S2P8C4 O76515 A0A212FN63 A0A212FN45 A0A1E1WC74 Q2QEH2 A0A0N1I4W9 I4DJD9 S4P8B4 Q0WX97 O61236 A0A0K8SYG9 B6CMG0 A0A2A4J1H5 I4DN38 M4M9L0 B3P4J3 Q6F440 H9JTT0 A0A212FN10 A0A0N0PCC3 A0A194R6W7 Q9VGM2 A0A3G1T1H0 Q8IGA2 A0A194Q0K6 A0A2W1BHB1 A0A0C9R4E4 A0A1I8M6S3 B4HI13 R4WCK5 B4PU50 A0A0A7BYM6 A0A2H1VTL2 A0A1I8MQH5 H9JTT3 A0A0T6BBK9 A0A1A9V645 A0A0K8SEJ6 I4DNC2 A0A162NXM9 A0A194PZX5 A0A194Q064 A0A0P5TIC9 A0A0P5XGE0 A0A0P6G6Z7 A0A2A4AQU9 A0A0P5M8L4 A0A1W4WC51 A0A0P5ZZS2 A0A0N8B910 A0A194R2V4 A0A0P5IYD3 A0A0P4ZX95 A0A0P5ATF9 A0A139WGC8 A0A0P6GQJ5 A0A0P5QXR5 A0A0P5L5I2 A0A0P5M8N3 A0A0N8B2G7 A0A0P5X8U2 A0A0P6GYC8 A0A0N8BW82 A0A2H1VWS6 A0A0P5Y9G7
B6CMF8 A0A2H1VZX6 P31417 A0A2A4IXI1 A0A2H1VZX7 A0A2H1V0J1 B6CMF9 A0A2A4IZR3 A0A2A4IZ24 A0A2W1BCW9 A0A2W1BK37 F8URE4 A0A2W1BCW8 A0A2A4IY62 A0A194Q1K0 A0A2H1VZW5 A0A2H1V1V6 A0A212FN08 A0A194R6U0 A0A2A4IYX0 A0A212FN12 Q2PWA4 A0A2H1W0N8 A0A194Q0Y2 A0A0N1IFX3 A0A2W1BHC4 A0A0N1I994 A0A1E1WE07 A0A3S2P8C4 O76515 A0A212FN63 A0A212FN45 A0A1E1WC74 Q2QEH2 A0A0N1I4W9 I4DJD9 S4P8B4 Q0WX97 O61236 A0A0K8SYG9 B6CMG0 A0A2A4J1H5 I4DN38 M4M9L0 B3P4J3 Q6F440 H9JTT0 A0A212FN10 A0A0N0PCC3 A0A194R6W7 Q9VGM2 A0A3G1T1H0 Q8IGA2 A0A194Q0K6 A0A2W1BHB1 A0A0C9R4E4 A0A1I8M6S3 B4HI13 R4WCK5 B4PU50 A0A0A7BYM6 A0A2H1VTL2 A0A1I8MQH5 H9JTT3 A0A0T6BBK9 A0A1A9V645 A0A0K8SEJ6 I4DNC2 A0A162NXM9 A0A194PZX5 A0A194Q064 A0A0P5TIC9 A0A0P5XGE0 A0A0P6G6Z7 A0A2A4AQU9 A0A0P5M8L4 A0A1W4WC51 A0A0P5ZZS2 A0A0N8B910 A0A194R2V4 A0A0P5IYD3 A0A0P4ZX95 A0A0P5ATF9 A0A139WGC8 A0A0P6GQJ5 A0A0P5QXR5 A0A0P5L5I2 A0A0P5M8N3 A0A0N8B2G7 A0A0P5X8U2 A0A0P6GYC8 A0A0N8BW82 A0A2H1VWS6 A0A0P5Y9G7
Pubmed
EMBL
BABH01003494
BABH01003501
DQ311357
AB262580
ABD36301.1
BAE96010.1
+ More
BABH01003497 M77754 KZ150128 PZC73091.1 NWSH01004822 PCG64669.1 EU325558 ACB54948.1 ODYU01005494 SOQ46401.1 M77755 PCG64667.1 SOQ46400.1 ODYU01000112 SOQ34331.1 EU325559 ACB54949.1 PZC73093.1 PCG64670.1 PCG64668.1 PZC73092.1 PZC73090.1 JF896323 AEH16743.1 PZC73089.1 PCG64671.1 KQ459582 KPI98869.1 SOQ46399.1 SOQ34332.1 AGBW02007648 OWR55132.1 KQ460878 KPJ11576.1 PCG64666.1 OWR55131.1 DQ299942 ABC02870.1 SOQ46402.1 KPI98973.1 KQ460635 KPJ13423.1 PZC73094.1 KPJ13424.1 GDQN01005887 GDQN01000306 JAT85167.1 JAT90748.1 RSAL01000205 RVE44355.1 AF074436 AAC25674.1 OWR55130.1 OWR55129.1 GDQN01006476 JAT84578.1 DQ109670 DQ311348 DQ539018 EF112401 AB262579 AAZ82096.1 ABD36292.1 ABG20590.1 ABK97427.1 BAE96009.1 KQ461183 KPJ07543.1 AK401407 BAM18029.1 KPI98807.1 GAIX01006016 JAA86544.1 AB267808 BAF02663.1 U75307 AAC24317.1 GBRD01007489 JAG58332.1 EU325560 KZ149898 ACB54950.1 PZC78673.1 NWSH01003893 PCG65719.1 AK402706 BAM19328.1 KC127672 AGG56524.1 CH954181 EDV49508.1 AB180450 BAD26694.1 BABH01003492 BABH01003493 AGBW02007650 OWR55117.1 KPJ13422.1 KPJ11606.1 AE014297 BT001757 BT001835 KX532042 AAN13512.1 AAN71512.1 AAN71590.1 ANY27852.1 MG992392 AXY94830.1 BT001880 AAN71654.1 KPI98838.1 PZC73095.1 GBYB01011169 JAG80936.1 CH480815 EDW42595.1 AK416948 BAN20163.1 CM000160 EDW97700.1 KC966936 AHV85210.1 ODYU01004372 SOQ44185.1 LJIG01002210 KRT84730.1 GBRD01014100 JAG51726.1 AK402833 BAM19412.1 LRGB01000512 KZS18353.1 KPI98871.1 KPI98972.1 GDIP01127413 JAL76301.1 GDIP01074122 JAM29593.1 GDIQ01048086 JAN46651.1 NWMT01000107 PCC99322.1 GDIQ01159142 JAK92583.1 GDIP01049618 JAM54097.1 GDIQ01200979 JAK50746.1 KPJ11575.1 GDIQ01209026 JAK42699.1 GDIP01207352 JAJ16050.1 GDIP01199099 JAJ24303.1 KQ971345 KYB27002.1 GDIQ01030122 JAN64615.1 GDIQ01117457 JAL34269.1 GDIQ01199678 JAK52047.1 GDIQ01159141 JAK92584.1 GDIQ01219323 JAK32402.1 GDIP01087762 JAM15953.1 GDIQ01048085 JAN46652.1 GDIQ01138803 JAL12923.1 ODYU01004915 SOQ45280.1 GDIP01062520 JAM41195.1
BABH01003497 M77754 KZ150128 PZC73091.1 NWSH01004822 PCG64669.1 EU325558 ACB54948.1 ODYU01005494 SOQ46401.1 M77755 PCG64667.1 SOQ46400.1 ODYU01000112 SOQ34331.1 EU325559 ACB54949.1 PZC73093.1 PCG64670.1 PCG64668.1 PZC73092.1 PZC73090.1 JF896323 AEH16743.1 PZC73089.1 PCG64671.1 KQ459582 KPI98869.1 SOQ46399.1 SOQ34332.1 AGBW02007648 OWR55132.1 KQ460878 KPJ11576.1 PCG64666.1 OWR55131.1 DQ299942 ABC02870.1 SOQ46402.1 KPI98973.1 KQ460635 KPJ13423.1 PZC73094.1 KPJ13424.1 GDQN01005887 GDQN01000306 JAT85167.1 JAT90748.1 RSAL01000205 RVE44355.1 AF074436 AAC25674.1 OWR55130.1 OWR55129.1 GDQN01006476 JAT84578.1 DQ109670 DQ311348 DQ539018 EF112401 AB262579 AAZ82096.1 ABD36292.1 ABG20590.1 ABK97427.1 BAE96009.1 KQ461183 KPJ07543.1 AK401407 BAM18029.1 KPI98807.1 GAIX01006016 JAA86544.1 AB267808 BAF02663.1 U75307 AAC24317.1 GBRD01007489 JAG58332.1 EU325560 KZ149898 ACB54950.1 PZC78673.1 NWSH01003893 PCG65719.1 AK402706 BAM19328.1 KC127672 AGG56524.1 CH954181 EDV49508.1 AB180450 BAD26694.1 BABH01003492 BABH01003493 AGBW02007650 OWR55117.1 KPJ13422.1 KPJ11606.1 AE014297 BT001757 BT001835 KX532042 AAN13512.1 AAN71512.1 AAN71590.1 ANY27852.1 MG992392 AXY94830.1 BT001880 AAN71654.1 KPI98838.1 PZC73095.1 GBYB01011169 JAG80936.1 CH480815 EDW42595.1 AK416948 BAN20163.1 CM000160 EDW97700.1 KC966936 AHV85210.1 ODYU01004372 SOQ44185.1 LJIG01002210 KRT84730.1 GBRD01014100 JAG51726.1 AK402833 BAM19412.1 LRGB01000512 KZS18353.1 KPI98871.1 KPI98972.1 GDIP01127413 JAL76301.1 GDIP01074122 JAM29593.1 GDIQ01048086 JAN46651.1 NWMT01000107 PCC99322.1 GDIQ01159142 JAK92583.1 GDIP01049618 JAM54097.1 GDIQ01200979 JAK50746.1 KPJ11575.1 GDIQ01209026 JAK42699.1 GDIP01207352 JAJ16050.1 GDIP01199099 JAJ24303.1 KQ971345 KYB27002.1 GDIQ01030122 JAN64615.1 GDIQ01117457 JAL34269.1 GDIQ01199678 JAK52047.1 GDIQ01159141 JAK92584.1 GDIQ01219323 JAK32402.1 GDIP01087762 JAM15953.1 GDIQ01048085 JAN46652.1 GDIQ01138803 JAL12923.1 ODYU01004915 SOQ45280.1 GDIP01062520 JAM41195.1
Proteomes
Pfam
PF00061 Lipocalin
Interpro
SUPFAM
SSF50814
SSF50814
Gene 3D
ProteinModelPortal
H9JTM8
Q2F5R6
H9JTT1
P31416
A0A2W1BFP9
A0A2A4IYF3
+ More
B6CMF8 A0A2H1VZX6 P31417 A0A2A4IXI1 A0A2H1VZX7 A0A2H1V0J1 B6CMF9 A0A2A4IZR3 A0A2A4IZ24 A0A2W1BCW9 A0A2W1BK37 F8URE4 A0A2W1BCW8 A0A2A4IY62 A0A194Q1K0 A0A2H1VZW5 A0A2H1V1V6 A0A212FN08 A0A194R6U0 A0A2A4IYX0 A0A212FN12 Q2PWA4 A0A2H1W0N8 A0A194Q0Y2 A0A0N1IFX3 A0A2W1BHC4 A0A0N1I994 A0A1E1WE07 A0A3S2P8C4 O76515 A0A212FN63 A0A212FN45 A0A1E1WC74 Q2QEH2 A0A0N1I4W9 I4DJD9 S4P8B4 Q0WX97 O61236 A0A0K8SYG9 B6CMG0 A0A2A4J1H5 I4DN38 M4M9L0 B3P4J3 Q6F440 H9JTT0 A0A212FN10 A0A0N0PCC3 A0A194R6W7 Q9VGM2 A0A3G1T1H0 Q8IGA2 A0A194Q0K6 A0A2W1BHB1 A0A0C9R4E4 A0A1I8M6S3 B4HI13 R4WCK5 B4PU50 A0A0A7BYM6 A0A2H1VTL2 A0A1I8MQH5 H9JTT3 A0A0T6BBK9 A0A1A9V645 A0A0K8SEJ6 I4DNC2 A0A162NXM9 A0A194PZX5 A0A194Q064 A0A0P5TIC9 A0A0P5XGE0 A0A0P6G6Z7 A0A2A4AQU9 A0A0P5M8L4 A0A1W4WC51 A0A0P5ZZS2 A0A0N8B910 A0A194R2V4 A0A0P5IYD3 A0A0P4ZX95 A0A0P5ATF9 A0A139WGC8 A0A0P6GQJ5 A0A0P5QXR5 A0A0P5L5I2 A0A0P5M8N3 A0A0N8B2G7 A0A0P5X8U2 A0A0P6GYC8 A0A0N8BW82 A0A2H1VWS6 A0A0P5Y9G7
B6CMF8 A0A2H1VZX6 P31417 A0A2A4IXI1 A0A2H1VZX7 A0A2H1V0J1 B6CMF9 A0A2A4IZR3 A0A2A4IZ24 A0A2W1BCW9 A0A2W1BK37 F8URE4 A0A2W1BCW8 A0A2A4IY62 A0A194Q1K0 A0A2H1VZW5 A0A2H1V1V6 A0A212FN08 A0A194R6U0 A0A2A4IYX0 A0A212FN12 Q2PWA4 A0A2H1W0N8 A0A194Q0Y2 A0A0N1IFX3 A0A2W1BHC4 A0A0N1I994 A0A1E1WE07 A0A3S2P8C4 O76515 A0A212FN63 A0A212FN45 A0A1E1WC74 Q2QEH2 A0A0N1I4W9 I4DJD9 S4P8B4 Q0WX97 O61236 A0A0K8SYG9 B6CMG0 A0A2A4J1H5 I4DN38 M4M9L0 B3P4J3 Q6F440 H9JTT0 A0A212FN10 A0A0N0PCC3 A0A194R6W7 Q9VGM2 A0A3G1T1H0 Q8IGA2 A0A194Q0K6 A0A2W1BHB1 A0A0C9R4E4 A0A1I8M6S3 B4HI13 R4WCK5 B4PU50 A0A0A7BYM6 A0A2H1VTL2 A0A1I8MQH5 H9JTT3 A0A0T6BBK9 A0A1A9V645 A0A0K8SEJ6 I4DNC2 A0A162NXM9 A0A194PZX5 A0A194Q064 A0A0P5TIC9 A0A0P5XGE0 A0A0P6G6Z7 A0A2A4AQU9 A0A0P5M8L4 A0A1W4WC51 A0A0P5ZZS2 A0A0N8B910 A0A194R2V4 A0A0P5IYD3 A0A0P4ZX95 A0A0P5ATF9 A0A139WGC8 A0A0P6GQJ5 A0A0P5QXR5 A0A0P5L5I2 A0A0P5M8N3 A0A0N8B2G7 A0A0P5X8U2 A0A0P6GYC8 A0A0N8BW82 A0A2H1VWS6 A0A0P5Y9G7
PDB
1MDC
E-value=2.51337e-29,
Score=314
Ontologies
KEGG
GO
PANTHER
Topology
Subcellular location
Cytoplasm
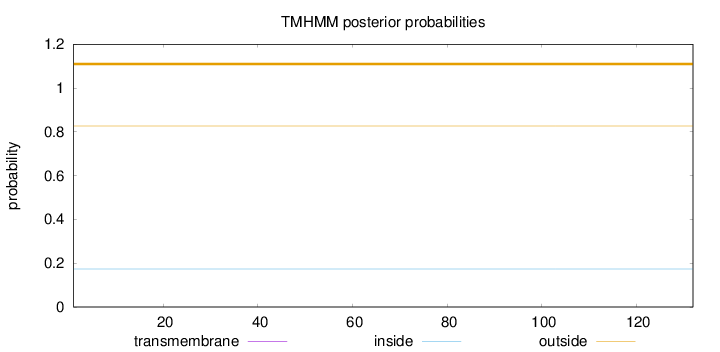
Length:
132
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.17375
outside
1 - 132
Population Genetic Test Statistics
Pi
154.547412
Theta
149.23951
Tajima's D
0.093655
CLR
138.005397
CSRT
0.388630568471576
Interpretation
Uncertain
