Pre Gene Modal
BGIBMGA012946
Annotation
PREDICTED:_probable_ATP-dependent_RNA_helicase_kurz_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 4.041
Sequence
CDS
ATGGGAAAGTCTAGATACAATAGTAAGGCACGACAAGTTGTAAAGACAAATATTGATGATTCAAAAACTAATGAGATTAAGTTAGAATTTGCAAAATCTGAATATGGAAATGCCGATGCCAGTAATGCACTGGCTTTGCCATCTAAGAAGAGGCAAACAAGGGTGGTTAAAGAGAAAATAGAGAAGACACATTTTCTGTCAAAAGCTCAAAGGAAAAGATTGGAGAAGATTGTTGATAAAAAAAAGAAAAAAGAAAATAGAGCTGCTCTATTAGAATCACTTTCACAAGTACAAGCAACTCCCGAGGAAGTGAAACAGTTAACAGCCATATCTGCAGTCCAAACAATAGGGTTGCGGAAACTTAATGAGTTCAGTGTCGAAGCTTTTATGACCGAAAGCTTTGATAAACCTGAGGTAACACAACACAAGAAGATCAGCAGTATTGCTGGTGCTAAAAAACGTTTACGTCTTCTGAAACTGGAGAAGTCCGAAAAGAAGAAATCAAAATACGATCCCAATGTGGTTGGATTGGAACTCGAAGAAGATGAAAATTCATCTGATGATTCTGACAGTACAGACGAAGTGACAAACACAATTGAAGATGGATCCCAAAATGTCATTGATATTACAGAAAGTATGACTAAAAATAATAATGAGGCCACCAACAACATTAATGAAATCAACAAAACTATAAAATTGGATAATACGAAAACAAGTACTACGCTACAGGAACAGAAAGAAGATAATATCACTGATAAATTAAAAGTAATCAGTAAAAGTGTTAAAGATAAAGTGAAAATAAAAAAACCTTTATTAGAACACCCCACAGTGCATGTTAATGTGAAAAGAGACCCTAAAGTGCAGGTAGCAAGACTCAAACTCCCAGTACTCGGCGAGGAACAGAGAATAATGGAGTTAGTGAATGAGAATCAATTTCTTATTGTTGCGGGTGAAACAGGTAGTGGAAAGACAACTCAGATACCACAGTTCCTCTATGAAGCGGGGTATGCAGAGAAGAAAATGGTAGCTGTAACAGAACCTAGGCGTGTCGCTACGGTAGCCATGTCTGCTAGAGTTGCTCATGAATTGGCTTTAACTAACAAAGAGGTCTCGTATCTAATGCGTTTTGAAGGTAACACTACGAAGGACACAAAAATTAAATTTATGACTGATGGTGTACTATTAAAGGAGATTCAATCAGATTTCTTATTAAGCAAGTATTCGGTTGTGATAATAGACGAGGCTCACGAGAGAAGCGTATACACAGATATACTCTTGGGTTTGTTGTCTAGAATAGTCCCGTTGAGGTGTAAACGAGGCAGTCCGCTTCGTTTAATCATTATGTCGGCTACATTGAGAGTTGAAGACTTTACGGAGAACACTAGATTGTTTAAACAGCCCCCGCCTGTTATAAAGATCGAATCAAGACAGTTCCCAGTTACGGTGCATTTTAACAAAAACACATACAAGGATTATTTGAAGGAAGCATTCAGGAAAACAGTAAAAATACATACAAGGTTGCCTGAGGGTGGAATACTTATATTTGTCACGGGGCAACAGGAGGTACGTTATCTACTCCGGAAATTGAAATCCTGTTTCCCGTACCGCAAAGATGTGGATTATTCAAAGTTGATCACTAAGAACACTAAGGGAGTAGAGGTTGACACTGCTCTCGATTCAGAACCAGATGACATCAACTCTGATGATGACGAGGTTGAGAAAGAGATGAAAAGGGCCCGCAAAGCCAGGAGGAAAGCGAAAAAGAAAGTTAAAGCTTTACCAAAGATCAATTTGGATGATTATGATATGCCTGAAGATGATGGTCAGGCTGATTTGGTGGGCAATGATGACGACAGCGAGGGGCACTTGAGTGATTCAGACGCAGAGGAAGACTCCCTGATGCCGGAAATAAAGTCGTGTCGTCAACCTCTATGGGTCTTGCCTCTGTACTCTATGCTAAACTCGAACAAGCAAGCTCGGGTCTTCGAACCTCCCCCGGTGGGCGCCAGGCTGTGCGTGGTCAGCACGGATGTCGCGGAGACGTCTCTAACGATACCGGCCATTAAATACGTTGTCGATACTGGAAAAAAGAAAATGAAGGTACACGACCACGTGACGGGCGCGAGCGCGTGGCGCGTTGTGTGGACGAGCCAGGCCAGCGCCGGGCAGCGGGCGGGGCGCGCGGGGCGGGTGTGCGCCGGCCACGTGTACCGGCTGTACAGCAGCGCGGTCTACCAGCATGACTGCCCGCCGCAGCACGCGCCTGACCTCTGCCGCAGGCCCGTCGACGACCTGGTCCTCATGATGAAGTGCATGGGCATCGATAAGGCACACGAATGCTTAACTGCCGTGGCCCGTCGGAAAGTTACTGGCTCGCTTCAAGGAAAACTATAA
Protein
MGKSRYNSKARQVVKTNIDDSKTNEIKLEFAKSEYGNADASNALALPSKKRQTRVVKEKIEKTHFLSKAQRKRLEKIVDKKKKKENRAALLESLSQVQATPEEVKQLTAISAVQTIGLRKLNEFSVEAFMTESFDKPEVTQHKKISSIAGAKKRLRLLKLEKSEKKKSKYDPNVVGLELEEDENSSDDSDSTDEVTNTIEDGSQNVIDITESMTKNNNEATNNINEINKTIKLDNTKTSTTLQEQKEDNITDKLKVISKSVKDKVKIKKPLLEHPTVHVNVKRDPKVQVARLKLPVLGEEQRIMELVNENQFLIVAGETGSGKTTQIPQFLYEAGYAEKKMVAVTEPRRVATVAMSARVAHELALTNKEVSYLMRFEGNTTKDTKIKFMTDGVLLKEIQSDFLLSKYSVVIIDEAHERSVYTDILLGLLSRIVPLRCKRGSPLRLIIMSATLRVEDFTENTRLFKQPPPVIKIESRQFPVTVHFNKNTYKDYLKEAFRKTVKIHTRLPEGGILIFVTGQQEVRYLLRKLKSCFPYRKDVDYSKLITKNTKGVEVDTALDSEPDDINSDDDEVEKEMKRARKARRKAKKKVKALPKINLDDYDMPEDDGQADLVGNDDDSEGHLSDSDAEEDSLMPEIKSCRQPLWVLPLYSMLNSNKQARVFEPPPVGARLCVVSTDVAETSLTIPAIKYVVDTGKKKMKVHDHVTGASAWRVVWTSQASAGQRAGRAGRVCAGHVYRLYSSAVYQHDCPPQHAPDLCRRPVDDLVLMMKCMGIDKAHECLTAVARRKVTGSLQGKL
Summary
Uniprot
Pubmed
EMBL
BABH01003502
BABH01003503
BABH01003504
KQ459582
KPI98867.1
AGBW02007648
+ More
OWR55134.1 RSAL01000100 RVE47545.1 GAMC01010992 JAB95563.1 JRES01000907 KNC27353.1 GAKP01020597 JAC38355.1 GAKP01020598 JAC38354.1 CH479205 EDW30760.1 NEVH01026085 PNF14952.1 KQ978957 KYN27205.1 ADTU01016112 GL888255 EGI63792.1 HACA01012955 CDW30316.1 ACPB03005725 KQ976725 KYM76540.1 KQ414670 KOC64378.1 GL448516 EFN84450.1
OWR55134.1 RSAL01000100 RVE47545.1 GAMC01010992 JAB95563.1 JRES01000907 KNC27353.1 GAKP01020597 JAC38355.1 GAKP01020598 JAC38354.1 CH479205 EDW30760.1 NEVH01026085 PNF14952.1 KQ978957 KYN27205.1 ADTU01016112 GL888255 EGI63792.1 HACA01012955 CDW30316.1 ACPB03005725 KQ976725 KYM76540.1 KQ414670 KOC64378.1 GL448516 EFN84450.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
PDB
6O16
E-value=4.5904e-137,
Score=1253
Ontologies
GO
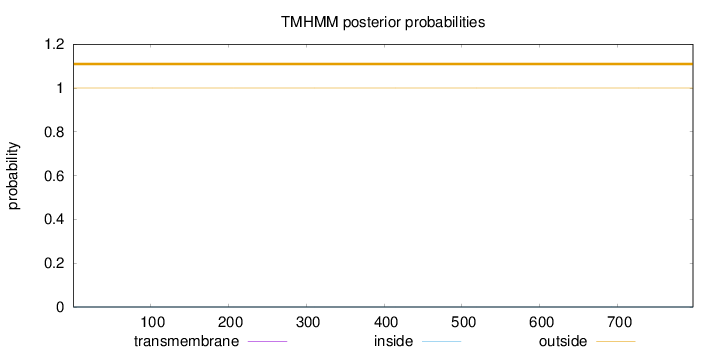
Topology
Length:
797
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00167
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00030
outside
1 - 797
Population Genetic Test Statistics
Pi
174.039091
Theta
150.885451
Tajima's D
0.20306
CLR
2.651196
CSRT
0.422828858557072
Interpretation
Uncertain
