Pre Gene Modal
BGIBMGA012946
Annotation
PREDICTED:_probable_ATP-dependent_RNA_helicase_kurz_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.289
Sequence
CDS
ATGAAGCCCCCAGATGAAAAGCAAGCCAGGCTGCTCAGGCAATTGGTGCTTAGTGGACTCGGAGATCAGGTCGGCAGGAAAATCGGCCTGGACGAGGTCAAAGAAGGCGAAGACAAAAGGAAATTCAAGTACGCTTACCACTGCGGCGACCTGGAGGAGCCCGTGCATCTGCACTCGGCTTCGGTGCTCCGCAAAACGATACCGGAGTGGGTGGTGTACCAGCAGCTATACGAGACGGGCGCCGAGGAACGAAGGAAGATGGTGATGAGCAACGTAACGGCCATCGAACCGGAATGGCTGCCCGTCTACGTGCCGCAGCTGTGCAATCTAGGTGAGCCTCTGTCTGATCCGGAGCCGCGTTACGAAGACGTAACTGGTAAAGTCCTATGTCACTACAAAGGCACGTTCGGCAAGGCGGGCTGGGAGCTGCCGGTTGTTGAAATCGAAATGCCCGAAAAAATTGAACGCTACCGATGGTTCGCGCGTTTCTTACTGGAGGGGTCAGTGTTTCCCAAACTTCGGAAGTACACGCCCTCTTTGTTGTCACCACCGTCGACGATGGTGAAATCGTGGGCGAAGCTGCAGCCCAGAACCGAGATATTATTAAAAGCTTTGTTAATGAAAAGAACCAGTACCAGGGAGTCGTTGAGGAAAATCTGGGAAGACCAGAAAAACTACTTACTTGAAGAGTATCTGAAGTGGCTTCCGGAGTCAGCCCATAACGAAGCTACATTGTACTGGCCGCCATTATGA
Protein
MKPPDEKQARLLRQLVLSGLGDQVGRKIGLDEVKEGEDKRKFKYAYHCGDLEEPVHLHSASVLRKTIPEWVVYQQLYETGAEERRKMVMSNVTAIEPEWLPVYVPQLCNLGEPLSDPEPRYEDVTGKVLCHYKGTFGKAGWELPVVEIEMPEKIERYRWFARFLLEGSVFPKLRKYTPSLLSPPSTMVKSWAKLQPRTEILLKALLMKRTSTRESLRKIWEDQKNYLLEEYLKWLPESAHNEATLYWPPL
Summary
Uniprot
H9JTT4
A0A3S2TJ77
A0A2A4JJ95
A0A2W1BIM9
A0A212FN09
S4PXL7
+ More
A0A194R1L0 A0A2J7PF36 A0A2J7PF51 A0A067RN61 A0A232EIX9 A0A3Q0JJX3 A0A154PJL3 A0A151WR00 A0A1J1HK52 A0A0J7NFQ4 A0A0L7R0L8 A0A087ZYB4 F4WPW3 E9IBM0 A0A195EG13 A0A195FDP0 A0A2A3EEH6 A0A1B6J8J0 A0A2P8XMH3 E2AIT2 A0A195AWW4 A0A195CKR7 A0A158NHY4 A0A1B6MBY0 N6U7L6 T1I963 A0A023F1U5 A0A224XH70 A0A1D2NA13 A0A0N8A389 A0A0P6B297 A0A162NF76 A0A0P4XV70 A0A0P6GSJ9 A0A0P5L590 A0A1B6CYI3 A0A0P5Q8S7 A0A0P6EEC9 A0A0P5GCZ0 A0A0P5IB35 A0A0P5NDC4 A0A0P5JDJ1 A0A0P6JZ06 E9G568 A0A0P6EKJ6 A0A1B6MT99 A0A1B6K7Y3 A0A146KWM9 A0A1B6FXZ6 A0A0A9YHQ9 A0A0P5XWM8 A0A0A9YDM0 T1J6J7 A0A226EPJ8 U4UB47 B4NE72 A0A0P6DZQ9 A0A026WX26 A0A3L8D9C7 A0A1A8SD33 A0A0T6B286 C3YFT9 D2A3P7 B4JWU3 B0W6U6 B4MA33 A0A0M4FBF8 B3MYT1 E2BIK3 R7UNM0 A0A1A8M5U1 K1Q1G9 V4AUC5 A0A0L0C4Q4 A0A2P4SKN3 B4L6V6 A0A3Q2EBR3 A0A3Q2EC87 A0A0P5F6Z6 A0A034V9B4 A0A034V771 A0A210Q0H9 A0A1Q3F0A1 A0A0K8VS77 F7F3D3 A0A1I8MU04 B3P8Z2 A0A1S3J317 A0A091TH09 B4R3G4 B4Q185 A0A1I8PKF2 A0A0A0A7D0 A0A093CV78
A0A194R1L0 A0A2J7PF36 A0A2J7PF51 A0A067RN61 A0A232EIX9 A0A3Q0JJX3 A0A154PJL3 A0A151WR00 A0A1J1HK52 A0A0J7NFQ4 A0A0L7R0L8 A0A087ZYB4 F4WPW3 E9IBM0 A0A195EG13 A0A195FDP0 A0A2A3EEH6 A0A1B6J8J0 A0A2P8XMH3 E2AIT2 A0A195AWW4 A0A195CKR7 A0A158NHY4 A0A1B6MBY0 N6U7L6 T1I963 A0A023F1U5 A0A224XH70 A0A1D2NA13 A0A0N8A389 A0A0P6B297 A0A162NF76 A0A0P4XV70 A0A0P6GSJ9 A0A0P5L590 A0A1B6CYI3 A0A0P5Q8S7 A0A0P6EEC9 A0A0P5GCZ0 A0A0P5IB35 A0A0P5NDC4 A0A0P5JDJ1 A0A0P6JZ06 E9G568 A0A0P6EKJ6 A0A1B6MT99 A0A1B6K7Y3 A0A146KWM9 A0A1B6FXZ6 A0A0A9YHQ9 A0A0P5XWM8 A0A0A9YDM0 T1J6J7 A0A226EPJ8 U4UB47 B4NE72 A0A0P6DZQ9 A0A026WX26 A0A3L8D9C7 A0A1A8SD33 A0A0T6B286 C3YFT9 D2A3P7 B4JWU3 B0W6U6 B4MA33 A0A0M4FBF8 B3MYT1 E2BIK3 R7UNM0 A0A1A8M5U1 K1Q1G9 V4AUC5 A0A0L0C4Q4 A0A2P4SKN3 B4L6V6 A0A3Q2EBR3 A0A3Q2EC87 A0A0P5F6Z6 A0A034V9B4 A0A034V771 A0A210Q0H9 A0A1Q3F0A1 A0A0K8VS77 F7F3D3 A0A1I8MU04 B3P8Z2 A0A1S3J317 A0A091TH09 B4R3G4 B4Q185 A0A1I8PKF2 A0A0A0A7D0 A0A093CV78
Pubmed
EMBL
BABH01003502
BABH01003503
BABH01003504
RSAL01000100
RVE47545.1
NWSH01001271
+ More
PCG71899.1 KZ150128 PZC73087.1 AGBW02007648 OWR55134.1 GAIX01004818 JAA87742.1 KQ460878 KPJ11577.1 NEVH01026085 PNF14950.1 PNF14952.1 KK852571 KDR21124.1 NNAY01004159 OXU18268.1 KQ434912 KZC11400.1 KQ982813 KYQ50329.1 CVRI01000005 CRK87788.1 LBMM01005595 KMQ91395.1 KQ414670 KOC64378.1 GL888255 EGI63792.1 GL762111 EFZ22036.1 KQ978957 KYN27205.1 KQ981636 KYN38780.1 KZ288266 PBC30118.1 GECU01012197 JAS95509.1 PYGN01001724 PSN33185.1 GL439878 EFN66658.1 KQ976725 KYM76540.1 KQ977622 KYN01320.1 ADTU01016112 GEBQ01006549 JAT33428.1 APGK01036710 KB740941 ENN77660.1 ACPB03005725 GBBI01003215 JAC15497.1 GFTR01008616 JAW07810.1 LJIJ01000125 ODN02093.1 GDIP01186825 JAJ36577.1 GDIP01023163 JAM80552.1 LRGB01000568 KZS17920.1 GDIP01237364 JAI86037.1 GDIQ01029358 JAN65379.1 GDIQ01181845 JAK69880.1 GEDC01018847 JAS18451.1 GDIQ01117615 JAL34111.1 GDIQ01063887 JAN30850.1 GDIQ01243928 GDIQ01242774 GDIQ01241329 GDIQ01239781 JAK08951.1 GDIQ01217135 JAK34590.1 GDIQ01144168 JAL07558.1 GDIQ01201638 JAK50087.1 GDIQ01002672 JAN92065.1 GL732532 EFX85421.1 GDIQ01074984 JAN19753.1 GEBQ01000849 JAT39128.1 GECU01000139 JAT07568.1 GDHC01019162 JAP99466.1 GECZ01014707 JAS55062.1 GBHO01012448 JAG31156.1 GDIP01066388 JAM37327.1 GBHO01012447 JAG31157.1 JH431878 LNIX01000003 OXA58751.1 KB632225 ERL90282.1 CH964239 EDW82041.1 GDIQ01069840 JAN24897.1 KK107078 EZA60291.1 QOIP01000011 RLU16508.1 HAEI01012981 SBS15450.1 LJIG01016144 KRT81484.1 GG666510 EEN60744.1 KQ971348 EFA05543.2 CH916376 EDV95219.1 DS231849 EDS36927.1 CH940655 EDW66092.1 CP012528 ALC49945.1 CH902632 EDV32775.1 GL448516 EFN84450.1 AMQN01001119 KB299619 ELU07825.1 HAEF01011228 SBR51896.1 JH818600 EKC30297.1 KB201305 ESO97341.1 JRES01000907 KNC27353.1 PPHD01039330 POI24662.1 CH933812 EDW06102.2 GDIQ01264783 JAJ86941.1 GAKP01020597 JAC38355.1 GAKP01020598 JAC38354.1 NEDP02005302 OWF42264.1 GFDL01014071 JAV20974.1 GDHF01010595 JAI41719.1 CH954183 EDV45597.1 KK472714 KFQ58051.1 CM000366 EDX16916.1 CM000162 EDX01392.1 KL870823 KGL89378.1 KL249182 KFV16824.1
PCG71899.1 KZ150128 PZC73087.1 AGBW02007648 OWR55134.1 GAIX01004818 JAA87742.1 KQ460878 KPJ11577.1 NEVH01026085 PNF14950.1 PNF14952.1 KK852571 KDR21124.1 NNAY01004159 OXU18268.1 KQ434912 KZC11400.1 KQ982813 KYQ50329.1 CVRI01000005 CRK87788.1 LBMM01005595 KMQ91395.1 KQ414670 KOC64378.1 GL888255 EGI63792.1 GL762111 EFZ22036.1 KQ978957 KYN27205.1 KQ981636 KYN38780.1 KZ288266 PBC30118.1 GECU01012197 JAS95509.1 PYGN01001724 PSN33185.1 GL439878 EFN66658.1 KQ976725 KYM76540.1 KQ977622 KYN01320.1 ADTU01016112 GEBQ01006549 JAT33428.1 APGK01036710 KB740941 ENN77660.1 ACPB03005725 GBBI01003215 JAC15497.1 GFTR01008616 JAW07810.1 LJIJ01000125 ODN02093.1 GDIP01186825 JAJ36577.1 GDIP01023163 JAM80552.1 LRGB01000568 KZS17920.1 GDIP01237364 JAI86037.1 GDIQ01029358 JAN65379.1 GDIQ01181845 JAK69880.1 GEDC01018847 JAS18451.1 GDIQ01117615 JAL34111.1 GDIQ01063887 JAN30850.1 GDIQ01243928 GDIQ01242774 GDIQ01241329 GDIQ01239781 JAK08951.1 GDIQ01217135 JAK34590.1 GDIQ01144168 JAL07558.1 GDIQ01201638 JAK50087.1 GDIQ01002672 JAN92065.1 GL732532 EFX85421.1 GDIQ01074984 JAN19753.1 GEBQ01000849 JAT39128.1 GECU01000139 JAT07568.1 GDHC01019162 JAP99466.1 GECZ01014707 JAS55062.1 GBHO01012448 JAG31156.1 GDIP01066388 JAM37327.1 GBHO01012447 JAG31157.1 JH431878 LNIX01000003 OXA58751.1 KB632225 ERL90282.1 CH964239 EDW82041.1 GDIQ01069840 JAN24897.1 KK107078 EZA60291.1 QOIP01000011 RLU16508.1 HAEI01012981 SBS15450.1 LJIG01016144 KRT81484.1 GG666510 EEN60744.1 KQ971348 EFA05543.2 CH916376 EDV95219.1 DS231849 EDS36927.1 CH940655 EDW66092.1 CP012528 ALC49945.1 CH902632 EDV32775.1 GL448516 EFN84450.1 AMQN01001119 KB299619 ELU07825.1 HAEF01011228 SBR51896.1 JH818600 EKC30297.1 KB201305 ESO97341.1 JRES01000907 KNC27353.1 PPHD01039330 POI24662.1 CH933812 EDW06102.2 GDIQ01264783 JAJ86941.1 GAKP01020597 JAC38355.1 GAKP01020598 JAC38354.1 NEDP02005302 OWF42264.1 GFDL01014071 JAV20974.1 GDHF01010595 JAI41719.1 CH954183 EDV45597.1 KK472714 KFQ58051.1 CM000366 EDX16916.1 CM000162 EDX01392.1 KL870823 KGL89378.1 KL249182 KFV16824.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053240
UP000235965
+ More
UP000027135 UP000215335 UP000079169 UP000076502 UP000075809 UP000183832 UP000036403 UP000053825 UP000005203 UP000007755 UP000078492 UP000078541 UP000242457 UP000245037 UP000000311 UP000078540 UP000078542 UP000005205 UP000019118 UP000015103 UP000094527 UP000076858 UP000000305 UP000198287 UP000030742 UP000007798 UP000053097 UP000279307 UP000001554 UP000007266 UP000001070 UP000002320 UP000008792 UP000092553 UP000007801 UP000008237 UP000014760 UP000005408 UP000030746 UP000037069 UP000009192 UP000265020 UP000242188 UP000002279 UP000095301 UP000008711 UP000085678 UP000000304 UP000002282 UP000095300 UP000053858
UP000027135 UP000215335 UP000079169 UP000076502 UP000075809 UP000183832 UP000036403 UP000053825 UP000005203 UP000007755 UP000078492 UP000078541 UP000242457 UP000245037 UP000000311 UP000078540 UP000078542 UP000005205 UP000019118 UP000015103 UP000094527 UP000076858 UP000000305 UP000198287 UP000030742 UP000007798 UP000053097 UP000279307 UP000001554 UP000007266 UP000001070 UP000002320 UP000008792 UP000092553 UP000007801 UP000008237 UP000014760 UP000005408 UP000030746 UP000037069 UP000009192 UP000265020 UP000242188 UP000002279 UP000095301 UP000008711 UP000085678 UP000000304 UP000002282 UP000095300 UP000053858
PRIDE
Pfam
Interpro
IPR007502
Helicase-assoc_dom
+ More
IPR027417 P-loop_NTPase
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR011545 DEAD/DEAH_box_helicase_dom
IPR011709 DUF1605
IPR001650 Helicase_C
IPR014001 Helicase_ATP-bd
IPR003593 AAA+_ATPase
IPR007303 TIP41-like
IPR007234 Vps53_N
IPR042035 DEAH_win-hel_dom
IPR000237 GRIP_dom
IPR012936 Erv_C
IPR039542 Erv_N
IPR021777 DUF3342
IPR027417 P-loop_NTPase
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR011545 DEAD/DEAH_box_helicase_dom
IPR011709 DUF1605
IPR001650 Helicase_C
IPR014001 Helicase_ATP-bd
IPR003593 AAA+_ATPase
IPR007303 TIP41-like
IPR007234 Vps53_N
IPR042035 DEAH_win-hel_dom
IPR000237 GRIP_dom
IPR012936 Erv_C
IPR039542 Erv_N
IPR021777 DUF3342
SUPFAM
SSF52540
SSF52540
Gene 3D
CDD
ProteinModelPortal
H9JTT4
A0A3S2TJ77
A0A2A4JJ95
A0A2W1BIM9
A0A212FN09
S4PXL7
+ More
A0A194R1L0 A0A2J7PF36 A0A2J7PF51 A0A067RN61 A0A232EIX9 A0A3Q0JJX3 A0A154PJL3 A0A151WR00 A0A1J1HK52 A0A0J7NFQ4 A0A0L7R0L8 A0A087ZYB4 F4WPW3 E9IBM0 A0A195EG13 A0A195FDP0 A0A2A3EEH6 A0A1B6J8J0 A0A2P8XMH3 E2AIT2 A0A195AWW4 A0A195CKR7 A0A158NHY4 A0A1B6MBY0 N6U7L6 T1I963 A0A023F1U5 A0A224XH70 A0A1D2NA13 A0A0N8A389 A0A0P6B297 A0A162NF76 A0A0P4XV70 A0A0P6GSJ9 A0A0P5L590 A0A1B6CYI3 A0A0P5Q8S7 A0A0P6EEC9 A0A0P5GCZ0 A0A0P5IB35 A0A0P5NDC4 A0A0P5JDJ1 A0A0P6JZ06 E9G568 A0A0P6EKJ6 A0A1B6MT99 A0A1B6K7Y3 A0A146KWM9 A0A1B6FXZ6 A0A0A9YHQ9 A0A0P5XWM8 A0A0A9YDM0 T1J6J7 A0A226EPJ8 U4UB47 B4NE72 A0A0P6DZQ9 A0A026WX26 A0A3L8D9C7 A0A1A8SD33 A0A0T6B286 C3YFT9 D2A3P7 B4JWU3 B0W6U6 B4MA33 A0A0M4FBF8 B3MYT1 E2BIK3 R7UNM0 A0A1A8M5U1 K1Q1G9 V4AUC5 A0A0L0C4Q4 A0A2P4SKN3 B4L6V6 A0A3Q2EBR3 A0A3Q2EC87 A0A0P5F6Z6 A0A034V9B4 A0A034V771 A0A210Q0H9 A0A1Q3F0A1 A0A0K8VS77 F7F3D3 A0A1I8MU04 B3P8Z2 A0A1S3J317 A0A091TH09 B4R3G4 B4Q185 A0A1I8PKF2 A0A0A0A7D0 A0A093CV78
A0A194R1L0 A0A2J7PF36 A0A2J7PF51 A0A067RN61 A0A232EIX9 A0A3Q0JJX3 A0A154PJL3 A0A151WR00 A0A1J1HK52 A0A0J7NFQ4 A0A0L7R0L8 A0A087ZYB4 F4WPW3 E9IBM0 A0A195EG13 A0A195FDP0 A0A2A3EEH6 A0A1B6J8J0 A0A2P8XMH3 E2AIT2 A0A195AWW4 A0A195CKR7 A0A158NHY4 A0A1B6MBY0 N6U7L6 T1I963 A0A023F1U5 A0A224XH70 A0A1D2NA13 A0A0N8A389 A0A0P6B297 A0A162NF76 A0A0P4XV70 A0A0P6GSJ9 A0A0P5L590 A0A1B6CYI3 A0A0P5Q8S7 A0A0P6EEC9 A0A0P5GCZ0 A0A0P5IB35 A0A0P5NDC4 A0A0P5JDJ1 A0A0P6JZ06 E9G568 A0A0P6EKJ6 A0A1B6MT99 A0A1B6K7Y3 A0A146KWM9 A0A1B6FXZ6 A0A0A9YHQ9 A0A0P5XWM8 A0A0A9YDM0 T1J6J7 A0A226EPJ8 U4UB47 B4NE72 A0A0P6DZQ9 A0A026WX26 A0A3L8D9C7 A0A1A8SD33 A0A0T6B286 C3YFT9 D2A3P7 B4JWU3 B0W6U6 B4MA33 A0A0M4FBF8 B3MYT1 E2BIK3 R7UNM0 A0A1A8M5U1 K1Q1G9 V4AUC5 A0A0L0C4Q4 A0A2P4SKN3 B4L6V6 A0A3Q2EBR3 A0A3Q2EC87 A0A0P5F6Z6 A0A034V9B4 A0A034V771 A0A210Q0H9 A0A1Q3F0A1 A0A0K8VS77 F7F3D3 A0A1I8MU04 B3P8Z2 A0A1S3J317 A0A091TH09 B4R3G4 B4Q185 A0A1I8PKF2 A0A0A0A7D0 A0A093CV78
PDB
6O16
E-value=4.01823e-59,
Score=575
Ontologies
GO
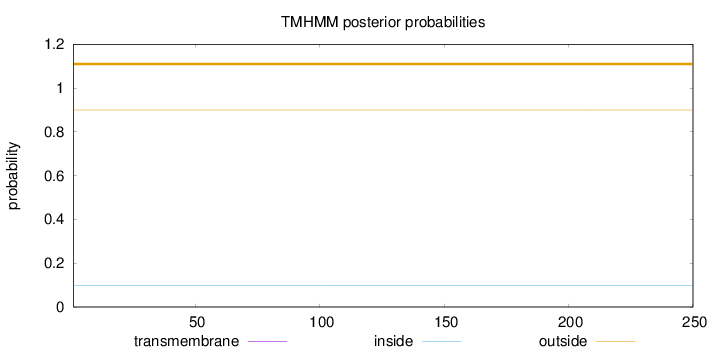
Topology
Length:
250
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000550000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.09975
outside
1 - 250
Population Genetic Test Statistics
Pi
221.822523
Theta
164.357375
Tajima's D
0.031292
CLR
141.636508
CSRT
0.380980950952452
Interpretation
Uncertain
