Gene
KWMTBOMO09714
Annotation
PREDICTED:_apyrase-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.113 PlasmaMembrane Reliability : 1.268
Sequence
CDS
ATGTATCTTATAATCGTTACGGTTAGTTTGTTTTTGGGTAATGTGTACAATTTCGTCTTACCCTTCGAAGGATTGTACAGATTGGATGTAATACATTACAACGACTTTCATGCGAGATTCGAGGAGACTTCGGTAAATACGCCGATATGCAAGTCTAATGATAGTGCCTGCCTGGGGGGGTTCCCTAGGCTCTACCACCACATCCAAACACTGCTAGTGGAAAAGCCACATTCAATACTCCTGAACGCGGGTGACAGTTTCCAGGGAACTTTCTGGTATACGCTGCTCAAATGGAACGTAACTCAAGAATTCATGAATATGCTACCTCACGACGCACATGCCATAGGTAACCATGAATTCGACGACGGGCCAGAAGGTCTTGCGCCATACTTATCTGCTCTTAATGCTCCTGTCGTGGCAGCCAATTTGGATGTTTCCAAAGAACCTTCGCTACAAAATCTAACCAAGCCTCATATCGTGATCGAACGACAAGGCAGGAAGATTGGAATCATTGGCTTGATAACGACTGATACCCAGATACTGTCCTTGCCCGGCAATGTGATTTTTACGGATCCATTCGAAGCCACGAGGCGGGAAACGGAAGCCTTGAATGAAAAAGGCGTGGATATCATCATACTTCTCTCACATTGCGGTCTTGAAGTTGACAAACAATTAGCTCGCGAACACGGAAAATACATCGATATAATAGTTGGAGGTCATTCGCATTCTCTTCTGTGGAATGGACCAGCGCCCAGTGGGGAGGAAGTCGCGGGGCCCTATCCAGTATTCGTCGAAAATTCTGCTGATGAAAAACACAAGGTATATAAATAA
Protein
MYLIIVTVSLFLGNVYNFVLPFEGLYRLDVIHYNDFHARFEETSVNTPICKSNDSACLGGFPRLYHHIQTLLVEKPHSILLNAGDSFQGTFWYTLLKWNVTQEFMNMLPHDAHAIGNHEFDDGPEGLAPYLSALNAPVVAANLDVSKEPSLQNLTKPHIVIERQGRKIGIIGLITTDTQILSLPGNVIFTDPFEATRRETEALNEKGVDIIILLSHCGLEVDKQLAREHGKYIDIIVGGHSHSLLWNGPAPSGEEVAGPYPVFVENSADEKHKVYK
Summary
Similarity
Belongs to the 5'-nucleotidase family.
Uniprot
A0A3G1T1Q5
A0A194R1A9
I4DS82
A0A212FN22
A0A2A4JIV7
A0A2H1VCG8
+ More
E0A922 A0A2W1BJS8 A0A437BAW0 A0A194R2W0 A0A194Q631 A0A194R238 A0A194Q1J5 A0A2A4JV45 A0A194PZX0 A0A2W1BHA1 A0A0N1IC27 A0A2H1WL38 A0A2H1VGJ2 H9JTT6 A0A3S2M1V1 H9JXD3 A0A437BB15 A0A212FN07 A0A2A4JVW4 A0A2A4JX59 A0A2A4JWR7 A0A2A4JW12 A0A194R1G7 A0A194R505 A0A212FMH9 A0A437B966 A0A067RQX4 A0A194PMT9 A0A2H1VIC8 A0A151IF43 A0A2H1WZX1 A0A182PGQ3 E2A7Z8 A0A151X5Z9 A0A026WZQ2 A0A3L8DU04 A0A1Y1N7H1 A0A195DPH7 A0A088AMD8 B5APR7 A0A0T6BA31 A0A151I0J3 A0A158NSU6 A0A182M997 A0A182VZA6 Q8T5H5 A0A182V0W0 A0A182HLB4 E2BRD8 A0A182L9P8 A0A182TN56 A0A310SIV8 A0A182R636 Q8MU74 A0A1J1J4F1 A0A182WSG4 A0A182YAL5 A0A2J7RI11 A0A139WMA1 W5JVI8 A0A3B0KAU5 B7S919 A0A2M4AV02 A0A182IVB6 A0A1W4WDV5 Q16UA0 H9JL90 A0A182K4G0 A0A084VT67 B4NEB2 A0A0Q9X374 U5EQR9 F4WSK6 A0A023ETZ8 A0A182GQ46 E9IPY5 A0A182J0X1 A0A182GQ45 A0A154P1M3 A0A182Q352 S6CVL9 A0A182NS57 A0A1Q3FN26 A0A182R0R0 D6WD64 V5G6C5 B0WJ00 U5EW72 A0A182JMY7 A0A1I8NCT3 A0A182U110 Q16QA5
E0A922 A0A2W1BJS8 A0A437BAW0 A0A194R2W0 A0A194Q631 A0A194R238 A0A194Q1J5 A0A2A4JV45 A0A194PZX0 A0A2W1BHA1 A0A0N1IC27 A0A2H1WL38 A0A2H1VGJ2 H9JTT6 A0A3S2M1V1 H9JXD3 A0A437BB15 A0A212FN07 A0A2A4JVW4 A0A2A4JX59 A0A2A4JWR7 A0A2A4JW12 A0A194R1G7 A0A194R505 A0A212FMH9 A0A437B966 A0A067RQX4 A0A194PMT9 A0A2H1VIC8 A0A151IF43 A0A2H1WZX1 A0A182PGQ3 E2A7Z8 A0A151X5Z9 A0A026WZQ2 A0A3L8DU04 A0A1Y1N7H1 A0A195DPH7 A0A088AMD8 B5APR7 A0A0T6BA31 A0A151I0J3 A0A158NSU6 A0A182M997 A0A182VZA6 Q8T5H5 A0A182V0W0 A0A182HLB4 E2BRD8 A0A182L9P8 A0A182TN56 A0A310SIV8 A0A182R636 Q8MU74 A0A1J1J4F1 A0A182WSG4 A0A182YAL5 A0A2J7RI11 A0A139WMA1 W5JVI8 A0A3B0KAU5 B7S919 A0A2M4AV02 A0A182IVB6 A0A1W4WDV5 Q16UA0 H9JL90 A0A182K4G0 A0A084VT67 B4NEB2 A0A0Q9X374 U5EQR9 F4WSK6 A0A023ETZ8 A0A182GQ46 E9IPY5 A0A182J0X1 A0A182GQ45 A0A154P1M3 A0A182Q352 S6CVL9 A0A182NS57 A0A1Q3FN26 A0A182R0R0 D6WD64 V5G6C5 B0WJ00 U5EW72 A0A182JMY7 A0A1I8NCT3 A0A182U110 Q16QA5
Pubmed
EMBL
MG770323
AXY94925.1
KQ460878
KPJ11578.1
AK405462
BAM20772.1
+ More
AGBW02007648 OWR55135.1 NWSH01001271 PCG71901.1 ODYU01001820 SOQ38540.1 HM569605 ADK90114.1 KZ150128 PZC73086.1 RSAL01000100 RVE47544.1 KPJ11580.1 KQ459582 KPI98865.1 KPJ11579.1 KPI98864.1 NWSH01000556 PCG75679.1 KPI98866.1 PZC73085.1 LADJ01006163 KPJ20897.1 ODYU01009397 SOQ53793.1 ODYU01002180 SOQ39344.1 BABH01003511 BABH01003512 RSAL01000067 RVE49304.1 BABH01041952 RVE47543.1 OWR55136.1 NWSH01000580 PCG75530.1 NWSH01000463 PCG76224.1 PCG76226.1 PCG76227.1 KPJ11542.1 KQ460685 KPJ12888.1 AGBW02007651 OWR54937.1 RSAL01000115 RVE46873.1 KK852475 KDR23050.1 KQ459600 KPI94308.1 ODYU01002721 SOQ40578.1 KQ977808 KYM99521.1 ODYU01012308 SOQ58557.1 GL437468 EFN70435.1 KQ982494 KYQ55710.1 KK107054 EZA61540.1 QOIP01000004 RLU23851.1 GEZM01010744 JAV93862.1 KQ980653 KYN14825.1 EU867772 ACF93470.1 LJIG01002991 KRT83949.1 KQ976619 KYM79119.1 ADTU01025187 ADTU01025188 AXCM01013925 AJ439398 AAAB01008987 CAD28126.1 EAA00943.2 APCN01000890 GL449962 EFN81716.1 KQ762882 OAD55322.1 AJ441131 CAD29633.1 CVRI01000070 CRL07285.1 NEVH01003506 PNF40456.1 KQ971312 KYB29062.1 ADMH02000310 ETN67025.1 OUUW01000035 SPP89842.1 EF710657 ACE75394.1 GGFK01011279 MBW44600.1 CH477627 EAT38116.1 BABH01011796 ATLV01016253 KE525074 KFB41161.1 CH964239 EDW82081.2 KRF99569.1 GANO01003160 JAB56711.1 GL888322 EGI62821.1 GAPW01001222 JAC12376.1 JXUM01079657 JXUM01079658 JXUM01079659 KQ563139 KXJ74403.1 GL764659 EFZ17310.1 JXUM01079656 KXJ74402.1 KQ434796 KZC05761.1 AXCN02000266 HF586473 CCQ71110.1 GFDL01006108 JAV28937.1 AXCN02000433 EEZ98343.1 GALX01002836 JAB65630.1 DS231954 EDS28861.1 GANO01000692 JAB59179.1 CH477755 EAT36583.1
AGBW02007648 OWR55135.1 NWSH01001271 PCG71901.1 ODYU01001820 SOQ38540.1 HM569605 ADK90114.1 KZ150128 PZC73086.1 RSAL01000100 RVE47544.1 KPJ11580.1 KQ459582 KPI98865.1 KPJ11579.1 KPI98864.1 NWSH01000556 PCG75679.1 KPI98866.1 PZC73085.1 LADJ01006163 KPJ20897.1 ODYU01009397 SOQ53793.1 ODYU01002180 SOQ39344.1 BABH01003511 BABH01003512 RSAL01000067 RVE49304.1 BABH01041952 RVE47543.1 OWR55136.1 NWSH01000580 PCG75530.1 NWSH01000463 PCG76224.1 PCG76226.1 PCG76227.1 KPJ11542.1 KQ460685 KPJ12888.1 AGBW02007651 OWR54937.1 RSAL01000115 RVE46873.1 KK852475 KDR23050.1 KQ459600 KPI94308.1 ODYU01002721 SOQ40578.1 KQ977808 KYM99521.1 ODYU01012308 SOQ58557.1 GL437468 EFN70435.1 KQ982494 KYQ55710.1 KK107054 EZA61540.1 QOIP01000004 RLU23851.1 GEZM01010744 JAV93862.1 KQ980653 KYN14825.1 EU867772 ACF93470.1 LJIG01002991 KRT83949.1 KQ976619 KYM79119.1 ADTU01025187 ADTU01025188 AXCM01013925 AJ439398 AAAB01008987 CAD28126.1 EAA00943.2 APCN01000890 GL449962 EFN81716.1 KQ762882 OAD55322.1 AJ441131 CAD29633.1 CVRI01000070 CRL07285.1 NEVH01003506 PNF40456.1 KQ971312 KYB29062.1 ADMH02000310 ETN67025.1 OUUW01000035 SPP89842.1 EF710657 ACE75394.1 GGFK01011279 MBW44600.1 CH477627 EAT38116.1 BABH01011796 ATLV01016253 KE525074 KFB41161.1 CH964239 EDW82081.2 KRF99569.1 GANO01003160 JAB56711.1 GL888322 EGI62821.1 GAPW01001222 JAC12376.1 JXUM01079657 JXUM01079658 JXUM01079659 KQ563139 KXJ74403.1 GL764659 EFZ17310.1 JXUM01079656 KXJ74402.1 KQ434796 KZC05761.1 AXCN02000266 HF586473 CCQ71110.1 GFDL01006108 JAV28937.1 AXCN02000433 EEZ98343.1 GALX01002836 JAB65630.1 DS231954 EDS28861.1 GANO01000692 JAB59179.1 CH477755 EAT36583.1
Proteomes
UP000053240
UP000007151
UP000218220
UP000283053
UP000053268
UP000005204
+ More
UP000027135 UP000078542 UP000075885 UP000000311 UP000075809 UP000053097 UP000279307 UP000078492 UP000005203 UP000078540 UP000005205 UP000075883 UP000075920 UP000007062 UP000075903 UP000075840 UP000008237 UP000075882 UP000075902 UP000075900 UP000183832 UP000076407 UP000076408 UP000235965 UP000007266 UP000000673 UP000268350 UP000075880 UP000192223 UP000008820 UP000075881 UP000030765 UP000007798 UP000007755 UP000069940 UP000249989 UP000076502 UP000075886 UP000075884 UP000002320 UP000095301
UP000027135 UP000078542 UP000075885 UP000000311 UP000075809 UP000053097 UP000279307 UP000078492 UP000005203 UP000078540 UP000005205 UP000075883 UP000075920 UP000007062 UP000075903 UP000075840 UP000008237 UP000075882 UP000075902 UP000075900 UP000183832 UP000076407 UP000076408 UP000235965 UP000007266 UP000000673 UP000268350 UP000075880 UP000192223 UP000008820 UP000075881 UP000030765 UP000007798 UP000007755 UP000069940 UP000249989 UP000076502 UP000075886 UP000075884 UP000002320 UP000095301
Interpro
SUPFAM
SSF55816
SSF55816
Gene 3D
ProteinModelPortal
A0A3G1T1Q5
A0A194R1A9
I4DS82
A0A212FN22
A0A2A4JIV7
A0A2H1VCG8
+ More
E0A922 A0A2W1BJS8 A0A437BAW0 A0A194R2W0 A0A194Q631 A0A194R238 A0A194Q1J5 A0A2A4JV45 A0A194PZX0 A0A2W1BHA1 A0A0N1IC27 A0A2H1WL38 A0A2H1VGJ2 H9JTT6 A0A3S2M1V1 H9JXD3 A0A437BB15 A0A212FN07 A0A2A4JVW4 A0A2A4JX59 A0A2A4JWR7 A0A2A4JW12 A0A194R1G7 A0A194R505 A0A212FMH9 A0A437B966 A0A067RQX4 A0A194PMT9 A0A2H1VIC8 A0A151IF43 A0A2H1WZX1 A0A182PGQ3 E2A7Z8 A0A151X5Z9 A0A026WZQ2 A0A3L8DU04 A0A1Y1N7H1 A0A195DPH7 A0A088AMD8 B5APR7 A0A0T6BA31 A0A151I0J3 A0A158NSU6 A0A182M997 A0A182VZA6 Q8T5H5 A0A182V0W0 A0A182HLB4 E2BRD8 A0A182L9P8 A0A182TN56 A0A310SIV8 A0A182R636 Q8MU74 A0A1J1J4F1 A0A182WSG4 A0A182YAL5 A0A2J7RI11 A0A139WMA1 W5JVI8 A0A3B0KAU5 B7S919 A0A2M4AV02 A0A182IVB6 A0A1W4WDV5 Q16UA0 H9JL90 A0A182K4G0 A0A084VT67 B4NEB2 A0A0Q9X374 U5EQR9 F4WSK6 A0A023ETZ8 A0A182GQ46 E9IPY5 A0A182J0X1 A0A182GQ45 A0A154P1M3 A0A182Q352 S6CVL9 A0A182NS57 A0A1Q3FN26 A0A182R0R0 D6WD64 V5G6C5 B0WJ00 U5EW72 A0A182JMY7 A0A1I8NCT3 A0A182U110 Q16QA5
E0A922 A0A2W1BJS8 A0A437BAW0 A0A194R2W0 A0A194Q631 A0A194R238 A0A194Q1J5 A0A2A4JV45 A0A194PZX0 A0A2W1BHA1 A0A0N1IC27 A0A2H1WL38 A0A2H1VGJ2 H9JTT6 A0A3S2M1V1 H9JXD3 A0A437BB15 A0A212FN07 A0A2A4JVW4 A0A2A4JX59 A0A2A4JWR7 A0A2A4JW12 A0A194R1G7 A0A194R505 A0A212FMH9 A0A437B966 A0A067RQX4 A0A194PMT9 A0A2H1VIC8 A0A151IF43 A0A2H1WZX1 A0A182PGQ3 E2A7Z8 A0A151X5Z9 A0A026WZQ2 A0A3L8DU04 A0A1Y1N7H1 A0A195DPH7 A0A088AMD8 B5APR7 A0A0T6BA31 A0A151I0J3 A0A158NSU6 A0A182M997 A0A182VZA6 Q8T5H5 A0A182V0W0 A0A182HLB4 E2BRD8 A0A182L9P8 A0A182TN56 A0A310SIV8 A0A182R636 Q8MU74 A0A1J1J4F1 A0A182WSG4 A0A182YAL5 A0A2J7RI11 A0A139WMA1 W5JVI8 A0A3B0KAU5 B7S919 A0A2M4AV02 A0A182IVB6 A0A1W4WDV5 Q16UA0 H9JL90 A0A182K4G0 A0A084VT67 B4NEB2 A0A0Q9X374 U5EQR9 F4WSK6 A0A023ETZ8 A0A182GQ46 E9IPY5 A0A182J0X1 A0A182GQ45 A0A154P1M3 A0A182Q352 S6CVL9 A0A182NS57 A0A1Q3FN26 A0A182R0R0 D6WD64 V5G6C5 B0WJ00 U5EW72 A0A182JMY7 A0A1I8NCT3 A0A182U110 Q16QA5
PDB
4H1S
E-value=1.99395e-40,
Score=415
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.610115
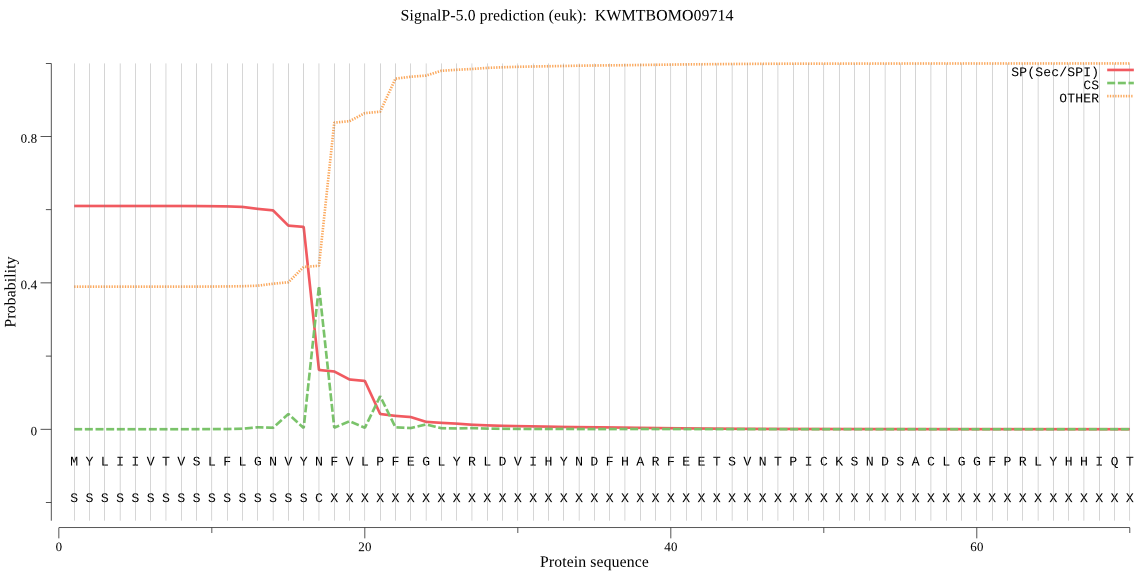
Length:
276
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
14.95708
Exp number, first 60 AAs:
14.66427
Total prob of N-in:
0.54776
POSSIBLE N-term signal
sequence
outside
1 - 276

Population Genetic Test Statistics
Pi
261.129113
Theta
17.082205
Tajima's D
0.677175
CLR
2.245754
CSRT
0.580520973951302
Interpretation
Uncertain
