Gene
KWMTBOMO09711 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014198
Annotation
PREDICTED:_apyrase-like_[Amyelois_transitella]
Full name
Apyrase
Alternative Name
ATP-diphosphatase
ATP-diphosphohydrolase
Adenosine diphosphatase
ATP-diphosphohydrolase
Adenosine diphosphatase
Location in the cell
Cytoplasmic Reliability : 2.79
Sequence
CDS
ATGATTGATTGGAACGGGGGCATGATCCGTTTGGACCGATCCATTCCTGAAGATCCAATGATGAAGGCAAAACTAAAACCATACATGAAACAAGTTCACGAAGGCGCAGCGGTGCCGATTGGTGAGACTCTAGTGGACATGCCGTTCAAGAATTGTCAAATTAAGGAGTGTGCGCTTGGAAATTTAGTAACAGACGCTTTTAATGAGAAAGCTATGAATACTGTCAAGACCGATTTGAGTTACATCGCATTCGCGCAATGTTCGTATCTTACTTCAAACATTCCAGAAGGAACTATCACCCACGGCCACATCGTCGAGCTGCTGCCTTACTACGATCAAATAGAAACATTCCAAATGAAAGGAAAATATATAATTGAGTTACTCGAGAGAAGCGCAAAGGATGTCCTACCATCAGAAATTCCGAATATGCTGCAAGTTTCTGGTTTGGAAATAGTTTATAATATGACTAAGCGGGAAGGAAAACGATTATACTCAGTGAAGGTCGGTCAAGAGAGCATGGACATCAACAAATTTTACCAGGTCACTGCCCCAGGATTTTTGGCTGACGGAGGTGATGGCTACACGATATTCAAAAAGCAGAAGAAAAACATGAAAGTTATCGGCTACGATCAAGACGTTTTAAAAGATTATATCAAGAAACATTCACCTATAGATTTAAATGTTGGTAACAGAATAAATATGGTCCGTTAG
Protein
MIDWNGGMIRLDRSIPEDPMMKAKLKPYMKQVHEGAAVPIGETLVDMPFKNCQIKECALGNLVTDAFNEKAMNTVKTDLSYIAFAQCSYLTSNIPEGTITHGHIVELLPYYDQIETFQMKGKYIIELLERSAKDVLPSEIPNMLQVSGLEIVYNMTKREGKRLYSVKVGQESMDINKFYQVTAPGFLADGGDGYTIFKKQKKNMKVIGYDQDVLKDYIKKHSPIDLNVGNRINMVR
Summary
Description
Facilitates hematophagy by inhibiting ADP-dependent platelet aggregation in the host. Shows potential for antithrombotic activity. May reduce probing time by facilitating the speed of locating blood.
Catalytic Activity
a ribonucleoside 5'-triphosphate + 2 H2O = a ribonucleoside 5'-phosphate + 2 H(+) + 2 phosphate
Cofactor
a divalent metal cation
Similarity
Belongs to the 5'-nucleotidase family.
Keywords
Allergen
ATP-binding
Direct protein sequencing
Hydrolase
Metal-binding
Nucleotide-binding
Secreted
Signal
Feature
chain Apyrase
Uniprot
H9JXD3
A0A3G1T1Q5
A0A437BAW0
A0A2H1VFF9
A0A2H1VCG8
A0A194PZX0
+ More
A0A2A4JIV7 A0A194R238 A0A212FN22 A0A194Q631 I4DS82 A0A2W1BJS8 E0A922 S4PC70 A0A212FN07 A0A2H1WL38 A0A2H1VGJ2 Q17NS8 A0A2W1BHA1 F1JZ10 B3A0N5 A0A0K8TQH7 A0A182GC77 Q9U9I6 A0A0L8HEY5 A0A1Q3FIZ4 A0A2G8JP94 A0A1L8DPP4 A0A1L8DPP0 A0A182JMY7 A0A1B0GP29 B0WJ00 A0A3S2M1V1 A0A182L9P8 Q8T5H5 A0A182HLB4 Q8MU74 A0A182WSG4 A0A194R6M3 A0A194Q1J5 A0A437BB15 A0A194R2W0 A0A182V0W0 A0A182TN56 A0A2G8LBN1 A0A0A1WD31 A0A1Y1N7H1 A0A182FA77 A0A1L8DPX0 H9JL89 A0A182K4G0 A0A084VT66 R7T630 A0A194PLJ4 A0A2A4JVW4 A0A151X5Z9 A0A336N6I4 W8BUB4 K1R448 A0A026WZQ2 A0A3L8DU04 W5J3U7 A0A336LDK1 A0A2M4AV02 D6WD64 A0A182PGQ3 W5J5F4 H9JL90 A0A1J1HLE0 A0A182FA78 A0A182NS57 Q95P65 A0A1B0ASU8 D3TRV7 W5J1N7 A0A182Q352 S6CVL9 E2A7Z8 A0A1I8NCS3 A0A194R505 A0A139WMA1 A0A182NMA1 J3JZ21 A0A1A9UUV1 H9JTT6 B7S919 A0A1A9XYG0 A0A2H1WZX1 A0A182STK5 A0A0N7ZAX7 K1R966 A0A336KUY8 A0A182J0X1 A0A336LSN5 A0A336LWJ0 A0A182Q7N3 A0A182R636 V5G6C5
A0A2A4JIV7 A0A194R238 A0A212FN22 A0A194Q631 I4DS82 A0A2W1BJS8 E0A922 S4PC70 A0A212FN07 A0A2H1WL38 A0A2H1VGJ2 Q17NS8 A0A2W1BHA1 F1JZ10 B3A0N5 A0A0K8TQH7 A0A182GC77 Q9U9I6 A0A0L8HEY5 A0A1Q3FIZ4 A0A2G8JP94 A0A1L8DPP4 A0A1L8DPP0 A0A182JMY7 A0A1B0GP29 B0WJ00 A0A3S2M1V1 A0A182L9P8 Q8T5H5 A0A182HLB4 Q8MU74 A0A182WSG4 A0A194R6M3 A0A194Q1J5 A0A437BB15 A0A194R2W0 A0A182V0W0 A0A182TN56 A0A2G8LBN1 A0A0A1WD31 A0A1Y1N7H1 A0A182FA77 A0A1L8DPX0 H9JL89 A0A182K4G0 A0A084VT66 R7T630 A0A194PLJ4 A0A2A4JVW4 A0A151X5Z9 A0A336N6I4 W8BUB4 K1R448 A0A026WZQ2 A0A3L8DU04 W5J3U7 A0A336LDK1 A0A2M4AV02 D6WD64 A0A182PGQ3 W5J5F4 H9JL90 A0A1J1HLE0 A0A182FA78 A0A182NS57 Q95P65 A0A1B0ASU8 D3TRV7 W5J1N7 A0A182Q352 S6CVL9 E2A7Z8 A0A1I8NCS3 A0A194R505 A0A139WMA1 A0A182NMA1 J3JZ21 A0A1A9UUV1 H9JTT6 B7S919 A0A1A9XYG0 A0A2H1WZX1 A0A182STK5 A0A0N7ZAX7 K1R966 A0A336KUY8 A0A182J0X1 A0A336LSN5 A0A336LWJ0 A0A182Q7N3 A0A182R636 V5G6C5
EC Number
3.6.1.5
Pubmed
19121390
26354079
22118469
22651552
28756777
22848670
+ More
23622113 17510324 21848516 26369729 26483478 10821849 29023486 20966253 12060762 12364791 14747013 17210077 25830018 28004739 24438588 23254933 24495485 22992520 24508170 30249741 20920257 23761445 18362917 19820115 20353571 23938757 20798317 25315136 22516182
23622113 17510324 21848516 26369729 26483478 10821849 29023486 20966253 12060762 12364791 14747013 17210077 25830018 28004739 24438588 23254933 24495485 22992520 24508170 30249741 20920257 23761445 18362917 19820115 20353571 23938757 20798317 25315136 22516182
EMBL
BABH01041952
MG770323
AXY94925.1
RSAL01000100
RVE47544.1
ODYU01002285
+ More
SOQ39567.1 ODYU01001820 SOQ38540.1 KQ459582 KPI98866.1 NWSH01001271 PCG71901.1 KQ460878 KPJ11579.1 AGBW02007648 OWR55135.1 KPI98865.1 AK405462 BAM20772.1 KZ150128 PZC73086.1 HM569605 ADK90114.1 GAIX01005602 JAA86958.1 OWR55136.1 ODYU01009397 SOQ53793.1 ODYU01002180 SOQ39344.1 CH477196 EAT48396.1 PZC73085.1 JF308210 ADX78255.1 GDAI01001190 JAI16413.1 JXUM01054688 KQ561832 KXJ77401.1 AF169229 AAD49730.1 KQ418431 KOF87350.1 GFDL01007465 JAV27580.1 MRZV01001480 PIK37567.1 GFDF01005652 JAV08432.1 GFDF01005651 JAV08433.1 AJVK01033050 DS231954 EDS28861.1 RSAL01000067 RVE49304.1 AJ439398 AAAB01008987 CAD28126.1 EAA00943.2 APCN01000890 AJ441131 CAD29633.1 KQ460685 KPJ12890.1 KPI98864.1 RVE47543.1 KPJ11580.1 MRZV01000135 PIK57676.1 GBXI01017413 JAC96878.1 GEZM01010744 JAV93862.1 GFDF01005657 JAV08427.1 BABH01011794 BABH01011795 BABH01011796 ATLV01016253 KE525074 KFB41160.1 AMQN01015158 KB311668 ELT88815.1 KQ459600 KPI94306.1 NWSH01000580 PCG75530.1 KQ982494 KYQ55710.1 UFQT01002888 SSX34218.1 GAMC01013841 JAB92714.1 JH823220 EKC28636.1 KK107054 EZA61540.1 QOIP01000004 RLU23851.1 ADMH02002183 ETN57988.1 UFQS01002906 UFQT01002906 SSX14867.1 SSX34255.1 GGFK01011279 MBW44600.1 KQ971312 EEZ98343.1 ETN57989.1 CVRI01000003 CRK87057.1 AF384674 AAK63848.1 JXJN01003008 EZ424159 ADD20435.1 ADMH02002165 ETN58132.1 AXCN02000266 HF586473 CCQ71110.1 GL437468 EFN70435.1 KPJ12888.1 KYB29062.1 BT128502 AEE63459.1 BABH01003511 BABH01003512 EF710657 ACE75394.1 ODYU01012308 SOQ58557.1 GDRN01092605 JAI60080.1 JH818488 EKC30511.1 UFQS01001030 UFQT01001030 SSX08629.1 SSX28545.1 UFQT01000158 SSX21032.1 SSX21033.1 AXCN02000938 GALX01002836 JAB65630.1
SOQ39567.1 ODYU01001820 SOQ38540.1 KQ459582 KPI98866.1 NWSH01001271 PCG71901.1 KQ460878 KPJ11579.1 AGBW02007648 OWR55135.1 KPI98865.1 AK405462 BAM20772.1 KZ150128 PZC73086.1 HM569605 ADK90114.1 GAIX01005602 JAA86958.1 OWR55136.1 ODYU01009397 SOQ53793.1 ODYU01002180 SOQ39344.1 CH477196 EAT48396.1 PZC73085.1 JF308210 ADX78255.1 GDAI01001190 JAI16413.1 JXUM01054688 KQ561832 KXJ77401.1 AF169229 AAD49730.1 KQ418431 KOF87350.1 GFDL01007465 JAV27580.1 MRZV01001480 PIK37567.1 GFDF01005652 JAV08432.1 GFDF01005651 JAV08433.1 AJVK01033050 DS231954 EDS28861.1 RSAL01000067 RVE49304.1 AJ439398 AAAB01008987 CAD28126.1 EAA00943.2 APCN01000890 AJ441131 CAD29633.1 KQ460685 KPJ12890.1 KPI98864.1 RVE47543.1 KPJ11580.1 MRZV01000135 PIK57676.1 GBXI01017413 JAC96878.1 GEZM01010744 JAV93862.1 GFDF01005657 JAV08427.1 BABH01011794 BABH01011795 BABH01011796 ATLV01016253 KE525074 KFB41160.1 AMQN01015158 KB311668 ELT88815.1 KQ459600 KPI94306.1 NWSH01000580 PCG75530.1 KQ982494 KYQ55710.1 UFQT01002888 SSX34218.1 GAMC01013841 JAB92714.1 JH823220 EKC28636.1 KK107054 EZA61540.1 QOIP01000004 RLU23851.1 ADMH02002183 ETN57988.1 UFQS01002906 UFQT01002906 SSX14867.1 SSX34255.1 GGFK01011279 MBW44600.1 KQ971312 EEZ98343.1 ETN57989.1 CVRI01000003 CRK87057.1 AF384674 AAK63848.1 JXJN01003008 EZ424159 ADD20435.1 ADMH02002165 ETN58132.1 AXCN02000266 HF586473 CCQ71110.1 GL437468 EFN70435.1 KPJ12888.1 KYB29062.1 BT128502 AEE63459.1 BABH01003511 BABH01003512 EF710657 ACE75394.1 ODYU01012308 SOQ58557.1 GDRN01092605 JAI60080.1 JH818488 EKC30511.1 UFQS01001030 UFQT01001030 SSX08629.1 SSX28545.1 UFQT01000158 SSX21032.1 SSX21033.1 AXCN02000938 GALX01002836 JAB65630.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000218220
UP000053240
UP000007151
+ More
UP000008820 UP000069940 UP000249989 UP000053454 UP000230750 UP000075880 UP000092462 UP000002320 UP000075882 UP000007062 UP000075840 UP000076407 UP000075903 UP000075902 UP000069272 UP000075881 UP000030765 UP000014760 UP000075809 UP000005408 UP000053097 UP000279307 UP000000673 UP000007266 UP000075885 UP000183832 UP000075884 UP000092460 UP000075886 UP000000311 UP000095301 UP000078200 UP000092443 UP000075901 UP000075900
UP000008820 UP000069940 UP000249989 UP000053454 UP000230750 UP000075880 UP000092462 UP000002320 UP000075882 UP000007062 UP000075840 UP000076407 UP000075903 UP000075902 UP000069272 UP000075881 UP000030765 UP000014760 UP000075809 UP000005408 UP000053097 UP000279307 UP000000673 UP000007266 UP000075885 UP000183832 UP000075884 UP000092460 UP000075886 UP000000311 UP000095301 UP000078200 UP000092443 UP000075901 UP000075900
Interpro
IPR036907
5'-Nucleotdase_C_sf
+ More
IPR008334 5'-Nucleotdase_C
IPR004843 Calcineurin-like_PHP_ApaH
IPR006179 5_nucleotidase/apyrase
IPR029052 Metallo-depent_PP-like
IPR006146 5'-Nucleotdase_CS
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013783 Ig-like_fold
IPR013098 Ig_I-set
IPR013106 Ig_V-set
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR008334 5'-Nucleotdase_C
IPR004843 Calcineurin-like_PHP_ApaH
IPR006179 5_nucleotidase/apyrase
IPR029052 Metallo-depent_PP-like
IPR006146 5'-Nucleotdase_CS
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013783 Ig-like_fold
IPR013098 Ig_I-set
IPR013106 Ig_V-set
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
Gene 3D
ProteinModelPortal
H9JXD3
A0A3G1T1Q5
A0A437BAW0
A0A2H1VFF9
A0A2H1VCG8
A0A194PZX0
+ More
A0A2A4JIV7 A0A194R238 A0A212FN22 A0A194Q631 I4DS82 A0A2W1BJS8 E0A922 S4PC70 A0A212FN07 A0A2H1WL38 A0A2H1VGJ2 Q17NS8 A0A2W1BHA1 F1JZ10 B3A0N5 A0A0K8TQH7 A0A182GC77 Q9U9I6 A0A0L8HEY5 A0A1Q3FIZ4 A0A2G8JP94 A0A1L8DPP4 A0A1L8DPP0 A0A182JMY7 A0A1B0GP29 B0WJ00 A0A3S2M1V1 A0A182L9P8 Q8T5H5 A0A182HLB4 Q8MU74 A0A182WSG4 A0A194R6M3 A0A194Q1J5 A0A437BB15 A0A194R2W0 A0A182V0W0 A0A182TN56 A0A2G8LBN1 A0A0A1WD31 A0A1Y1N7H1 A0A182FA77 A0A1L8DPX0 H9JL89 A0A182K4G0 A0A084VT66 R7T630 A0A194PLJ4 A0A2A4JVW4 A0A151X5Z9 A0A336N6I4 W8BUB4 K1R448 A0A026WZQ2 A0A3L8DU04 W5J3U7 A0A336LDK1 A0A2M4AV02 D6WD64 A0A182PGQ3 W5J5F4 H9JL90 A0A1J1HLE0 A0A182FA78 A0A182NS57 Q95P65 A0A1B0ASU8 D3TRV7 W5J1N7 A0A182Q352 S6CVL9 E2A7Z8 A0A1I8NCS3 A0A194R505 A0A139WMA1 A0A182NMA1 J3JZ21 A0A1A9UUV1 H9JTT6 B7S919 A0A1A9XYG0 A0A2H1WZX1 A0A182STK5 A0A0N7ZAX7 K1R966 A0A336KUY8 A0A182J0X1 A0A336LSN5 A0A336LWJ0 A0A182Q7N3 A0A182R636 V5G6C5
A0A2A4JIV7 A0A194R238 A0A212FN22 A0A194Q631 I4DS82 A0A2W1BJS8 E0A922 S4PC70 A0A212FN07 A0A2H1WL38 A0A2H1VGJ2 Q17NS8 A0A2W1BHA1 F1JZ10 B3A0N5 A0A0K8TQH7 A0A182GC77 Q9U9I6 A0A0L8HEY5 A0A1Q3FIZ4 A0A2G8JP94 A0A1L8DPP4 A0A1L8DPP0 A0A182JMY7 A0A1B0GP29 B0WJ00 A0A3S2M1V1 A0A182L9P8 Q8T5H5 A0A182HLB4 Q8MU74 A0A182WSG4 A0A194R6M3 A0A194Q1J5 A0A437BB15 A0A194R2W0 A0A182V0W0 A0A182TN56 A0A2G8LBN1 A0A0A1WD31 A0A1Y1N7H1 A0A182FA77 A0A1L8DPX0 H9JL89 A0A182K4G0 A0A084VT66 R7T630 A0A194PLJ4 A0A2A4JVW4 A0A151X5Z9 A0A336N6I4 W8BUB4 K1R448 A0A026WZQ2 A0A3L8DU04 W5J3U7 A0A336LDK1 A0A2M4AV02 D6WD64 A0A182PGQ3 W5J5F4 H9JL90 A0A1J1HLE0 A0A182FA78 A0A182NS57 Q95P65 A0A1B0ASU8 D3TRV7 W5J1N7 A0A182Q352 S6CVL9 E2A7Z8 A0A1I8NCS3 A0A194R505 A0A139WMA1 A0A182NMA1 J3JZ21 A0A1A9UUV1 H9JTT6 B7S919 A0A1A9XYG0 A0A2H1WZX1 A0A182STK5 A0A0N7ZAX7 K1R966 A0A336KUY8 A0A182J0X1 A0A336LSN5 A0A336LWJ0 A0A182Q7N3 A0A182R636 V5G6C5
PDB
4H2B
E-value=7.0771e-20,
Score=237
Ontologies
GO
PANTHER
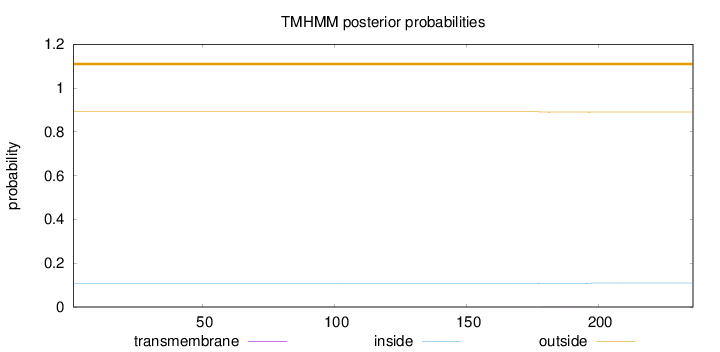
Topology
Subcellular location
Secreted
Length:
236
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04055
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.10844
outside
1 - 236
Population Genetic Test Statistics
Pi
428.859859
Theta
185.806312
Tajima's D
4.641006
CLR
0.579072
CSRT
0.99990000499975
Interpretation
Uncertain
