Gene
KWMTBOMO09709 Validated by peptides from experiments
Annotation
CRAL-TRIO_domain-containing_protein?_partial_[Manduca_sexta]
Location in the cell
PlasmaMembrane Reliability : 1.978
Sequence
CDS
ATGGCAAGAGCATACTTGGAGAACACGATAATAGTCAACAAAGGATCCATAGAAAGAGCCAAAGCCCAGATTAGTAGGCTTTGCGCATTGAAATCATTGTTGCCTACTTTCTTTGATGGCTACAAGTCCCGAAGCGATTTGCTGGAAACGAAAAAATACCTTTTACAAGCCTGGTTGCCGAGAATGACAGAGGAAAATCACAGGGTCTACGTAGTTAGAGTTAGTAATGGCGCAACGACGAAGTGTATTCTAAACGTCTACAAATTTGGCATTCTGCTAAACGAGTATTTACGAGCTCACGATTACAATGACGGTGTAATAGTGTTTATCGACTTCAGGAATGTAAACGTCATGGAGTTCGCTAGTAATTTGAATTTAGCGGATCTACGCAATATCATGACAATAGCGATGGAAGGATACGATGTGAGGGTTAAAGGTATACATTTCTTCACTTCATCGAAAACAATTGATGTGATTGTCAATATCATGAAACAGGTTGTTAGTGCCAAACTAGGGAGTCGCATCCACACGCACCTTACTTTAGATAGCCTTTACAAGTACATAACACCGAATGATATATTACCAGCTGAGTATGGCGGGAACGAAGAATCATTGGACGATTTAAGCGATAAATTAATTAACACCTTTACGAATGAAGATAATGTTAAATTCTTTGCTGAGATGGCCGAGGCTGGTGTTCATGAAGCTTTGAGGCCAAAGGATGGGGTGGCCTGTTTCGATGAACAATCTCTTGGTATAGCTGGAAGTTTTAGGACATTAAGTGTAGATTAA
Protein
MARAYLENTIIVNKGSIERAKAQISRLCALKSLLPTFFDGYKSRSDLLETKKYLLQAWLPRMTEENHRVYVVRVSNGATTKCILNVYKFGILLNEYLRAHDYNDGVIVFIDFRNVNVMEFASNLNLADLRNIMTIAMEGYDVRVKGIHFFTSSKTIDVIVNIMKQVVSAKLGSRIHTHLTLDSLYKYITPNDILPAEYGGNEESLDDLSDKLINTFTNEDNVKFFAEMAEAGVHEALRPKDGVACFDEQSLGIAGSFRTLSVD
Summary
Uniprot
A0A0S2Z393
A0A2W1BF53
A0A2H1VU69
A0A2A4J6B5
A0A0S2Z354
A0A212FN16
+ More
A0A212FN73 A0A194Q0M6 A0A2H4RMR5 A0A0S2Z340 A0A194PZV7 A0A2A4J610 A0A2H4RMR6 A0A194R1M2 A0A2W1BJJ9 A0A2H4RMR8 A0A2H4RMP8 A0A2H4RMR2 A0A194PZV1 A0A0S2Z358 A0A2A4JNW0 A0A194R247 A0A2H4RMS3 A0A2H4RMS9 A0A2H4RMT2 A0A2H4RMU5 A0A2H1UZW6 A0A3S2L7M8 A0A2H4RMT7 A0A2H1W0Z0 A0A3S2LZD4 A0A2A4JNK7 A0A212FN55 A0A2W1BI70 A0A2A4J4G2 A0A2H4RMU0 A0A437BAW6 A0A2H1W264 A0A2H4RMT4 A0A2H1WD56 A0A2W1BCD2 A0A2A4IUI0 A0A2H1W3J3 A0A2W1BML1 A0A2A4IW24 A0A2H1VXT5 A0A2H1V3H4 H9JKR5 A0A1E1WBH5 A0A0S2Z362 H9JKR6 A0A212FN46 A0A2H1VYX3 A0A0S2Z343 A0A2W1BCA0 A0A3S2NSP3 A0A2H1V516 A0A194R1B9 A0A2W1BGX0 A0A3S2PCJ5 A0A3S2NCI1 A0A1E1WFQ9 A0A2A4J2F6 A0A2W1BJA3 A0A2H1WDV2 A0A2A4J0W3 A0A2H1VM17 A0A0S2Z2V1 A0A2H4RMS8 A0A1E1VYT1 A0A2A4JMX0 A0A2W1BGS3 A0A2H4RMP7 A0A1Y1NHE0 D2A5R3 I4DIV6 A0A194Q6G5 A0A232EQE5 A0A194R1R7 A0A0S2Z3C6 I4DMP5 A0A3S2NT85 A0A2H1WHW8 A0A2H4RMS7 A0A2J7Q1W1 A0A2A4JN64 A0A2W1BQL9 A0A2H4RMV1 A0A195ECA1 V5I956
A0A212FN73 A0A194Q0M6 A0A2H4RMR5 A0A0S2Z340 A0A194PZV7 A0A2A4J610 A0A2H4RMR6 A0A194R1M2 A0A2W1BJJ9 A0A2H4RMR8 A0A2H4RMP8 A0A2H4RMR2 A0A194PZV1 A0A0S2Z358 A0A2A4JNW0 A0A194R247 A0A2H4RMS3 A0A2H4RMS9 A0A2H4RMT2 A0A2H4RMU5 A0A2H1UZW6 A0A3S2L7M8 A0A2H4RMT7 A0A2H1W0Z0 A0A3S2LZD4 A0A2A4JNK7 A0A212FN55 A0A2W1BI70 A0A2A4J4G2 A0A2H4RMU0 A0A437BAW6 A0A2H1W264 A0A2H4RMT4 A0A2H1WD56 A0A2W1BCD2 A0A2A4IUI0 A0A2H1W3J3 A0A2W1BML1 A0A2A4IW24 A0A2H1VXT5 A0A2H1V3H4 H9JKR5 A0A1E1WBH5 A0A0S2Z362 H9JKR6 A0A212FN46 A0A2H1VYX3 A0A0S2Z343 A0A2W1BCA0 A0A3S2NSP3 A0A2H1V516 A0A194R1B9 A0A2W1BGX0 A0A3S2PCJ5 A0A3S2NCI1 A0A1E1WFQ9 A0A2A4J2F6 A0A2W1BJA3 A0A2H1WDV2 A0A2A4J0W3 A0A2H1VM17 A0A0S2Z2V1 A0A2H4RMS8 A0A1E1VYT1 A0A2A4JMX0 A0A2W1BGS3 A0A2H4RMP7 A0A1Y1NHE0 D2A5R3 I4DIV6 A0A194Q6G5 A0A232EQE5 A0A194R1R7 A0A0S2Z3C6 I4DMP5 A0A3S2NT85 A0A2H1WHW8 A0A2H4RMS7 A0A2J7Q1W1 A0A2A4JN64 A0A2W1BQL9 A0A2H4RMV1 A0A195ECA1 V5I956
Pubmed
EMBL
KT943562
ALQ33320.1
KZ150143
PZC72921.1
ODYU01004475
SOQ44379.1
+ More
NWSH01003092 PCG66963.1 KT943558 ALQ33316.1 AGBW02007648 OWR55142.1 OWR55140.1 KQ459582 KPI98858.1 MG434624 ATY51930.1 KT943544 ALQ33302.1 KPI98856.1 PCG66964.1 MG434626 ATY51932.1 KQ460878 KPJ11587.1 PZC72920.1 MG434628 ATY51934.1 MG434605 ATY51911.1 MG434625 ATY51931.1 KPI98857.1 KT943545 ALQ33303.1 NWSH01000951 PCG73398.1 KPJ11589.1 MG434627 ATY51933.1 MG434629 ATY51935.1 MG434631 ATY51937.1 MG434650 ATY51956.1 ODYU01000041 SOQ34143.1 RSAL01000100 RVE47537.1 MG434643 ATY51949.1 ODYU01005386 SOQ46174.1 RVE47535.1 PCG73399.1 OWR55139.1 PZC72917.1 PCG66965.1 MG434648 ATY51954.1 RVE47538.1 ODYU01005865 SOQ47185.1 MG434630 ATY51936.1 ODYU01007545 SOQ50404.1 PZC72922.1 NWSH01006612 PCG63329.1 ODYU01006075 SOQ47608.1 PZC72923.1 PCG63330.1 ODYU01005115 SOQ45661.1 ODYU01000500 SOQ35381.1 BABH01011652 GDQN01006712 GDQN01000539 JAT84342.1 JAT90515.1 KT943537 ALQ33295.1 BABH01011650 BABH01011651 OWR55143.1 ODYU01005307 SOQ46031.1 KT943542 ALQ33300.1 PZC72919.1 RVE47536.1 ODYU01000690 SOQ35889.1 KPJ11588.1 PZC72924.1 RVE47533.1 RVE47534.1 GDQN01005214 GDQN01001800 JAT85840.1 JAT89254.1 NWSH01003956 PCG65620.1 PZC72916.1 ODYU01008002 SOQ51248.1 PCG65621.1 ODYU01003303 SOQ41879.1 KT943555 ALQ33313.1 MG434632 ATY51938.1 GDQN01011167 JAT79887.1 PCG73397.1 PZC72915.1 MG434603 ATY51909.1 GEZM01002435 JAV97261.1 KQ971345 EFA05407.1 AK401224 BAM17846.1 KPI98995.1 NNAY01002773 OXU20585.1 KPJ11637.1 KT943552 ALQ33310.1 AK402563 BAM19185.1 RSAL01000093 RVE47935.1 ODYU01008754 SOQ52627.1 MG434633 ATY51939.1 NEVH01019379 PNF22549.1 NWSH01001060 PCG72852.1 KZ150073 PZC74013.1 MG434656 ATY51962.1 KQ979074 KYN22850.1 GALX01003520 JAB64946.1
NWSH01003092 PCG66963.1 KT943558 ALQ33316.1 AGBW02007648 OWR55142.1 OWR55140.1 KQ459582 KPI98858.1 MG434624 ATY51930.1 KT943544 ALQ33302.1 KPI98856.1 PCG66964.1 MG434626 ATY51932.1 KQ460878 KPJ11587.1 PZC72920.1 MG434628 ATY51934.1 MG434605 ATY51911.1 MG434625 ATY51931.1 KPI98857.1 KT943545 ALQ33303.1 NWSH01000951 PCG73398.1 KPJ11589.1 MG434627 ATY51933.1 MG434629 ATY51935.1 MG434631 ATY51937.1 MG434650 ATY51956.1 ODYU01000041 SOQ34143.1 RSAL01000100 RVE47537.1 MG434643 ATY51949.1 ODYU01005386 SOQ46174.1 RVE47535.1 PCG73399.1 OWR55139.1 PZC72917.1 PCG66965.1 MG434648 ATY51954.1 RVE47538.1 ODYU01005865 SOQ47185.1 MG434630 ATY51936.1 ODYU01007545 SOQ50404.1 PZC72922.1 NWSH01006612 PCG63329.1 ODYU01006075 SOQ47608.1 PZC72923.1 PCG63330.1 ODYU01005115 SOQ45661.1 ODYU01000500 SOQ35381.1 BABH01011652 GDQN01006712 GDQN01000539 JAT84342.1 JAT90515.1 KT943537 ALQ33295.1 BABH01011650 BABH01011651 OWR55143.1 ODYU01005307 SOQ46031.1 KT943542 ALQ33300.1 PZC72919.1 RVE47536.1 ODYU01000690 SOQ35889.1 KPJ11588.1 PZC72924.1 RVE47533.1 RVE47534.1 GDQN01005214 GDQN01001800 JAT85840.1 JAT89254.1 NWSH01003956 PCG65620.1 PZC72916.1 ODYU01008002 SOQ51248.1 PCG65621.1 ODYU01003303 SOQ41879.1 KT943555 ALQ33313.1 MG434632 ATY51938.1 GDQN01011167 JAT79887.1 PCG73397.1 PZC72915.1 MG434603 ATY51909.1 GEZM01002435 JAV97261.1 KQ971345 EFA05407.1 AK401224 BAM17846.1 KPI98995.1 NNAY01002773 OXU20585.1 KPJ11637.1 KT943552 ALQ33310.1 AK402563 BAM19185.1 RSAL01000093 RVE47935.1 ODYU01008754 SOQ52627.1 MG434633 ATY51939.1 NEVH01019379 PNF22549.1 NWSH01001060 PCG72852.1 KZ150073 PZC74013.1 MG434656 ATY51962.1 KQ979074 KYN22850.1 GALX01003520 JAB64946.1
Proteomes
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A0S2Z393
A0A2W1BF53
A0A2H1VU69
A0A2A4J6B5
A0A0S2Z354
A0A212FN16
+ More
A0A212FN73 A0A194Q0M6 A0A2H4RMR5 A0A0S2Z340 A0A194PZV7 A0A2A4J610 A0A2H4RMR6 A0A194R1M2 A0A2W1BJJ9 A0A2H4RMR8 A0A2H4RMP8 A0A2H4RMR2 A0A194PZV1 A0A0S2Z358 A0A2A4JNW0 A0A194R247 A0A2H4RMS3 A0A2H4RMS9 A0A2H4RMT2 A0A2H4RMU5 A0A2H1UZW6 A0A3S2L7M8 A0A2H4RMT7 A0A2H1W0Z0 A0A3S2LZD4 A0A2A4JNK7 A0A212FN55 A0A2W1BI70 A0A2A4J4G2 A0A2H4RMU0 A0A437BAW6 A0A2H1W264 A0A2H4RMT4 A0A2H1WD56 A0A2W1BCD2 A0A2A4IUI0 A0A2H1W3J3 A0A2W1BML1 A0A2A4IW24 A0A2H1VXT5 A0A2H1V3H4 H9JKR5 A0A1E1WBH5 A0A0S2Z362 H9JKR6 A0A212FN46 A0A2H1VYX3 A0A0S2Z343 A0A2W1BCA0 A0A3S2NSP3 A0A2H1V516 A0A194R1B9 A0A2W1BGX0 A0A3S2PCJ5 A0A3S2NCI1 A0A1E1WFQ9 A0A2A4J2F6 A0A2W1BJA3 A0A2H1WDV2 A0A2A4J0W3 A0A2H1VM17 A0A0S2Z2V1 A0A2H4RMS8 A0A1E1VYT1 A0A2A4JMX0 A0A2W1BGS3 A0A2H4RMP7 A0A1Y1NHE0 D2A5R3 I4DIV6 A0A194Q6G5 A0A232EQE5 A0A194R1R7 A0A0S2Z3C6 I4DMP5 A0A3S2NT85 A0A2H1WHW8 A0A2H4RMS7 A0A2J7Q1W1 A0A2A4JN64 A0A2W1BQL9 A0A2H4RMV1 A0A195ECA1 V5I956
A0A212FN73 A0A194Q0M6 A0A2H4RMR5 A0A0S2Z340 A0A194PZV7 A0A2A4J610 A0A2H4RMR6 A0A194R1M2 A0A2W1BJJ9 A0A2H4RMR8 A0A2H4RMP8 A0A2H4RMR2 A0A194PZV1 A0A0S2Z358 A0A2A4JNW0 A0A194R247 A0A2H4RMS3 A0A2H4RMS9 A0A2H4RMT2 A0A2H4RMU5 A0A2H1UZW6 A0A3S2L7M8 A0A2H4RMT7 A0A2H1W0Z0 A0A3S2LZD4 A0A2A4JNK7 A0A212FN55 A0A2W1BI70 A0A2A4J4G2 A0A2H4RMU0 A0A437BAW6 A0A2H1W264 A0A2H4RMT4 A0A2H1WD56 A0A2W1BCD2 A0A2A4IUI0 A0A2H1W3J3 A0A2W1BML1 A0A2A4IW24 A0A2H1VXT5 A0A2H1V3H4 H9JKR5 A0A1E1WBH5 A0A0S2Z362 H9JKR6 A0A212FN46 A0A2H1VYX3 A0A0S2Z343 A0A2W1BCA0 A0A3S2NSP3 A0A2H1V516 A0A194R1B9 A0A2W1BGX0 A0A3S2PCJ5 A0A3S2NCI1 A0A1E1WFQ9 A0A2A4J2F6 A0A2W1BJA3 A0A2H1WDV2 A0A2A4J0W3 A0A2H1VM17 A0A0S2Z2V1 A0A2H4RMS8 A0A1E1VYT1 A0A2A4JMX0 A0A2W1BGS3 A0A2H4RMP7 A0A1Y1NHE0 D2A5R3 I4DIV6 A0A194Q6G5 A0A232EQE5 A0A194R1R7 A0A0S2Z3C6 I4DMP5 A0A3S2NT85 A0A2H1WHW8 A0A2H4RMS7 A0A2J7Q1W1 A0A2A4JN64 A0A2W1BQL9 A0A2H4RMV1 A0A195ECA1 V5I956
PDB
1R5L
E-value=1.08636e-05,
Score=115
Ontologies
GO
Topology
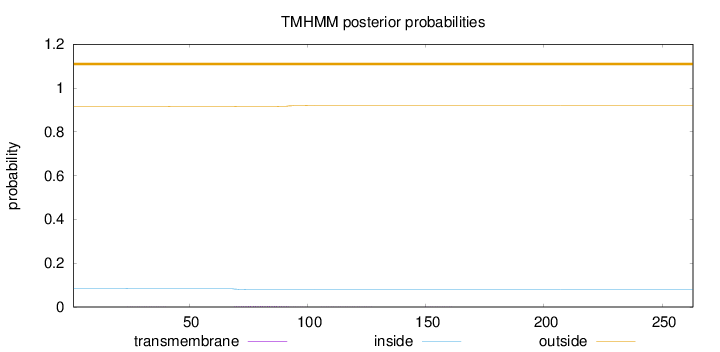
Length:
263
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.09177
Exp number, first 60 AAs:
0.00274
Total prob of N-in:
0.08381
outside
1 - 263
Population Genetic Test Statistics
Pi
302.355235
Theta
174.867844
Tajima's D
3.124302
CLR
0.29098
CSRT
0.984250787460627
Interpretation
Uncertain
