Gene
KWMTBOMO09706
Pre Gene Modal
BGIBMGA012949
Annotation
PREDICTED:_potassium_channel_subfamily_K_member_18-like_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.296 PlasmaMembrane Reliability : 1.136
Sequence
CDS
ATGGACAAAAACAAGAAAAAGAACCATTTTAATGGAAAATATAAGAAAAATGGCAAAAACGAGGCTTACAAAACGGAAGACGATATCATCACTACGTGCTGTCTCTGCGTTAAAACAAGAAAGGGGAAAAAGAAGAGTTTGATCGCTGGTTGTGTCACTAATTTGGGTATTTTTGTTTTACTTCTAGCGTACACGTTGTTGGGCGCGTTTATCTTCTTAGCTATAGAAGGGAATTCTTCAAAAATCCATCAGAAAACACTCGCGACCACATCTTATCAAGTTAACGATAAAAATTCAGTGCCCTCAACAAAGTTGAACAGTAGTATAAGTCAAGCTAGCGCGGAACTTCGATCACAGACGGTCGAAAGTATTTGGGAAATCACAGTTTCATTAAACATACTTTACAAAGAGAACTGGACTAGACTGGCAGCACAAGAGATAGCAAGATTTCAAGAGAAGTTAGTAGCTAGAGTCGCAGCTGATGTTTCAGCTCAGTATGGAGGAGCAAGAGCACTGGAATCAGCACCAGATTTGGTTGTTGACGACTATGAATGGAATTTCGCGAAAGCTTTCCTGTATTCATTGACCGTTTTAACAACTATAGGCTACGGCAGCGTTGCACCACGTACGGCACTTGGTGAATCTTTACCTGAAGTTTTGCACCATATAGGTTCAAGATATGATGGCGGAGCTATTCGCCGGACAGTATACACTCTTAGAGAGGAGAAAGAGCCAGAGCGTGTGACCCCTATGTAA
Protein
MDKNKKKNHFNGKYKKNGKNEAYKTEDDIITTCCLCVKTRKGKKKSLIAGCVTNLGIFVLLLAYTLLGAFIFLAIEGNSSKIHQKTLATTSYQVNDKNSVPSTKLNSSISQASAELRSQTVESIWEITVSLNILYKENWTRLAAQEIARFQEKLVARVAADVSAQYGGARALESAPDLVVDDYEWNFAKAFLYSLTVLTTIGYGSVAPRTALGESLPEVLHHIGSRYDGGAIRRTVYTLREEKEPERVTPM
Summary
Similarity
Belongs to the two pore domain potassium channel (TC 1.A.1.8) family.
Uniprot
H9JTT7
A0A3S2NY61
A0A2H1VRM0
A0A2W1BHB4
A0A194R6U4
A0A2A4K337
+ More
A0A0L7KMZ7 A0A194Q0N1 A0A212FN26 A0A1L8DET1 A0A2H1X3T4 A0A1J1ID08 A0A0L7L7U6 A0A2A4JM27 A0A2W1BAR8 A0A0N0U782 A0A182PAA6 D2A6I4 A0A154PFT0 A0A182IMD3 H9JTM0 E2BG43 A0A212FN32 A0A182GXF6 A0A182LFS8 A0A1B0GH38 A0A3S2LIQ5 A0A194R1M7 A0A158NJU8 A0A1S4HDT8 A0A182UQN6 A0A182MLI1 A0A182W079 A0A182WRI1 A0A194Q1I5 B0X5L6 A0A182HU96 A0A195C5I4 A0A3L8DE05 A0A182RNP6 A0A310SJZ9 E9ITV9 A0A026WT28 A0A1B6EDW4 A0A195B675 A0A182N624 A0A195EKK5 A0A182Y4C5 W5JF99 A0A182F7F2 E2AV51 A0A151XJY9 K7IQ88 A0A1W5CAD0 A0A182U5S3 F4WCC8 A0A182Q2A8 A0A1B6JU73 A0A084VRE0 A0A2J7RCY2 A0A067R7L9 A0A2R7WN62 A0A182K1R4 A0A2H1W8J8 A0A2S2NWI9 J9JQB1 A0A2H8TI30 A0A0N1I6X3 E0VY73 A0A182GEM1 B4M7Y1 A0A182NN86 A0A0L7RCA3 A0A0L0C7T3 A0A336KTM2 A0A182WAT8 A0A182PNG0
A0A0L7KMZ7 A0A194Q0N1 A0A212FN26 A0A1L8DET1 A0A2H1X3T4 A0A1J1ID08 A0A0L7L7U6 A0A2A4JM27 A0A2W1BAR8 A0A0N0U782 A0A182PAA6 D2A6I4 A0A154PFT0 A0A182IMD3 H9JTM0 E2BG43 A0A212FN32 A0A182GXF6 A0A182LFS8 A0A1B0GH38 A0A3S2LIQ5 A0A194R1M7 A0A158NJU8 A0A1S4HDT8 A0A182UQN6 A0A182MLI1 A0A182W079 A0A182WRI1 A0A194Q1I5 B0X5L6 A0A182HU96 A0A195C5I4 A0A3L8DE05 A0A182RNP6 A0A310SJZ9 E9ITV9 A0A026WT28 A0A1B6EDW4 A0A195B675 A0A182N624 A0A195EKK5 A0A182Y4C5 W5JF99 A0A182F7F2 E2AV51 A0A151XJY9 K7IQ88 A0A1W5CAD0 A0A182U5S3 F4WCC8 A0A182Q2A8 A0A1B6JU73 A0A084VRE0 A0A2J7RCY2 A0A067R7L9 A0A2R7WN62 A0A182K1R4 A0A2H1W8J8 A0A2S2NWI9 J9JQB1 A0A2H8TI30 A0A0N1I6X3 E0VY73 A0A182GEM1 B4M7Y1 A0A182NN86 A0A0L7RCA3 A0A0L0C7T3 A0A336KTM2 A0A182WAT8 A0A182PNG0
Pubmed
EMBL
BABH01003515
RSAL01000100
RVE47542.1
ODYU01004008
SOQ43447.1
KZ150128
+ More
PZC73084.1 KQ460878 KPJ11581.1 NWSH01000202 PCG78459.1 JTDY01008277 KOB64648.1 KQ459582 KPI98863.1 AGBW02007648 OWR55137.1 GFDF01009136 JAV04948.1 ODYU01013224 SOQ59918.1 CVRI01000047 CRK98167.1 JTDY01002366 KOB71567.1 NWSH01001064 PCG72836.1 KZ150267 PZC71701.1 KQ435709 KOX79958.1 KQ971348 EFA04915.1 KQ434893 KZC10672.1 BABH01003557 GL448115 EFN85311.1 OWR55145.1 JXUM01095279 JXUM01095280 JXUM01095281 JXUM01095282 KQ564185 KXJ72611.1 AJWK01003950 RVE47532.1 KPJ11592.1 ADTU01001990 ADTU01001991 ADTU01001992 ADTU01001993 ADTU01001994 ADTU01001995 AAAB01008859 AXCM01001083 KPI98854.1 DS232386 EDS40953.1 APCN01000613 KQ978292 KYM95433.1 QOIP01000009 RLU18665.1 KQ764395 OAD54526.1 GL765726 EFZ16000.1 KK107119 EZA58836.1 GEDC01001176 JAS36122.1 KQ976582 KYM79767.1 KQ978739 KYN28800.1 ADMH02001371 ETN62761.1 GL443026 EFN62682.1 KQ982052 KYQ60651.1 EGK96654.1 GL888070 EGI68108.1 AXCN02000193 GECU01005253 JAT02454.1 ATLV01015606 KE525023 KFB40534.1 NEVH01005366 PNF38691.1 KK852648 KDR19509.1 KK855092 PTY20781.1 ODYU01006903 SOQ49172.1 GGMR01008467 MBY21086.1 ABLF02035901 ABLF02035904 ABLF02035907 ABLF02035909 ABLF02035910 GFXV01001982 MBW13787.1 KQ460652 KPJ12973.1 DS235844 EEB18329.1 JXUM01058168 JXUM01058169 JXUM01058170 JXUM01058171 JXUM01058172 JXUM01058173 KQ561990 KXJ76972.1 CH940653 EDW62898.2 KQ414616 KOC68463.1 JRES01000767 KNC28458.1 UFQS01001015 UFQT01001015 SSX08586.1 SSX28502.1
PZC73084.1 KQ460878 KPJ11581.1 NWSH01000202 PCG78459.1 JTDY01008277 KOB64648.1 KQ459582 KPI98863.1 AGBW02007648 OWR55137.1 GFDF01009136 JAV04948.1 ODYU01013224 SOQ59918.1 CVRI01000047 CRK98167.1 JTDY01002366 KOB71567.1 NWSH01001064 PCG72836.1 KZ150267 PZC71701.1 KQ435709 KOX79958.1 KQ971348 EFA04915.1 KQ434893 KZC10672.1 BABH01003557 GL448115 EFN85311.1 OWR55145.1 JXUM01095279 JXUM01095280 JXUM01095281 JXUM01095282 KQ564185 KXJ72611.1 AJWK01003950 RVE47532.1 KPJ11592.1 ADTU01001990 ADTU01001991 ADTU01001992 ADTU01001993 ADTU01001994 ADTU01001995 AAAB01008859 AXCM01001083 KPI98854.1 DS232386 EDS40953.1 APCN01000613 KQ978292 KYM95433.1 QOIP01000009 RLU18665.1 KQ764395 OAD54526.1 GL765726 EFZ16000.1 KK107119 EZA58836.1 GEDC01001176 JAS36122.1 KQ976582 KYM79767.1 KQ978739 KYN28800.1 ADMH02001371 ETN62761.1 GL443026 EFN62682.1 KQ982052 KYQ60651.1 EGK96654.1 GL888070 EGI68108.1 AXCN02000193 GECU01005253 JAT02454.1 ATLV01015606 KE525023 KFB40534.1 NEVH01005366 PNF38691.1 KK852648 KDR19509.1 KK855092 PTY20781.1 ODYU01006903 SOQ49172.1 GGMR01008467 MBY21086.1 ABLF02035901 ABLF02035904 ABLF02035907 ABLF02035909 ABLF02035910 GFXV01001982 MBW13787.1 KQ460652 KPJ12973.1 DS235844 EEB18329.1 JXUM01058168 JXUM01058169 JXUM01058170 JXUM01058171 JXUM01058172 JXUM01058173 KQ561990 KXJ76972.1 CH940653 EDW62898.2 KQ414616 KOC68463.1 JRES01000767 KNC28458.1 UFQS01001015 UFQT01001015 SSX08586.1 SSX28502.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000218220
UP000037510
UP000053268
+ More
UP000007151 UP000183832 UP000053105 UP000075885 UP000007266 UP000076502 UP000075880 UP000008237 UP000069940 UP000249989 UP000075882 UP000092461 UP000005205 UP000075903 UP000075883 UP000075920 UP000076407 UP000002320 UP000075840 UP000078542 UP000279307 UP000075900 UP000053097 UP000078540 UP000075884 UP000078492 UP000076408 UP000000673 UP000069272 UP000000311 UP000075809 UP000002358 UP000007062 UP000075902 UP000007755 UP000075886 UP000030765 UP000235965 UP000027135 UP000075881 UP000007819 UP000009046 UP000008792 UP000053825 UP000037069
UP000007151 UP000183832 UP000053105 UP000075885 UP000007266 UP000076502 UP000075880 UP000008237 UP000069940 UP000249989 UP000075882 UP000092461 UP000005205 UP000075903 UP000075883 UP000075920 UP000076407 UP000002320 UP000075840 UP000078542 UP000279307 UP000075900 UP000053097 UP000078540 UP000075884 UP000078492 UP000076408 UP000000673 UP000069272 UP000000311 UP000075809 UP000002358 UP000007062 UP000075902 UP000007755 UP000075886 UP000030765 UP000235965 UP000027135 UP000075881 UP000007819 UP000009046 UP000008792 UP000053825 UP000037069
Pfam
PF07885 Ion_trans_2
ProteinModelPortal
H9JTT7
A0A3S2NY61
A0A2H1VRM0
A0A2W1BHB4
A0A194R6U4
A0A2A4K337
+ More
A0A0L7KMZ7 A0A194Q0N1 A0A212FN26 A0A1L8DET1 A0A2H1X3T4 A0A1J1ID08 A0A0L7L7U6 A0A2A4JM27 A0A2W1BAR8 A0A0N0U782 A0A182PAA6 D2A6I4 A0A154PFT0 A0A182IMD3 H9JTM0 E2BG43 A0A212FN32 A0A182GXF6 A0A182LFS8 A0A1B0GH38 A0A3S2LIQ5 A0A194R1M7 A0A158NJU8 A0A1S4HDT8 A0A182UQN6 A0A182MLI1 A0A182W079 A0A182WRI1 A0A194Q1I5 B0X5L6 A0A182HU96 A0A195C5I4 A0A3L8DE05 A0A182RNP6 A0A310SJZ9 E9ITV9 A0A026WT28 A0A1B6EDW4 A0A195B675 A0A182N624 A0A195EKK5 A0A182Y4C5 W5JF99 A0A182F7F2 E2AV51 A0A151XJY9 K7IQ88 A0A1W5CAD0 A0A182U5S3 F4WCC8 A0A182Q2A8 A0A1B6JU73 A0A084VRE0 A0A2J7RCY2 A0A067R7L9 A0A2R7WN62 A0A182K1R4 A0A2H1W8J8 A0A2S2NWI9 J9JQB1 A0A2H8TI30 A0A0N1I6X3 E0VY73 A0A182GEM1 B4M7Y1 A0A182NN86 A0A0L7RCA3 A0A0L0C7T3 A0A336KTM2 A0A182WAT8 A0A182PNG0
A0A0L7KMZ7 A0A194Q0N1 A0A212FN26 A0A1L8DET1 A0A2H1X3T4 A0A1J1ID08 A0A0L7L7U6 A0A2A4JM27 A0A2W1BAR8 A0A0N0U782 A0A182PAA6 D2A6I4 A0A154PFT0 A0A182IMD3 H9JTM0 E2BG43 A0A212FN32 A0A182GXF6 A0A182LFS8 A0A1B0GH38 A0A3S2LIQ5 A0A194R1M7 A0A158NJU8 A0A1S4HDT8 A0A182UQN6 A0A182MLI1 A0A182W079 A0A182WRI1 A0A194Q1I5 B0X5L6 A0A182HU96 A0A195C5I4 A0A3L8DE05 A0A182RNP6 A0A310SJZ9 E9ITV9 A0A026WT28 A0A1B6EDW4 A0A195B675 A0A182N624 A0A195EKK5 A0A182Y4C5 W5JF99 A0A182F7F2 E2AV51 A0A151XJY9 K7IQ88 A0A1W5CAD0 A0A182U5S3 F4WCC8 A0A182Q2A8 A0A1B6JU73 A0A084VRE0 A0A2J7RCY2 A0A067R7L9 A0A2R7WN62 A0A182K1R4 A0A2H1W8J8 A0A2S2NWI9 J9JQB1 A0A2H8TI30 A0A0N1I6X3 E0VY73 A0A182GEM1 B4M7Y1 A0A182NN86 A0A0L7RCA3 A0A0L0C7T3 A0A336KTM2 A0A182WAT8 A0A182PNG0
PDB
4RUE
E-value=0.00121629,
Score=97
Ontologies
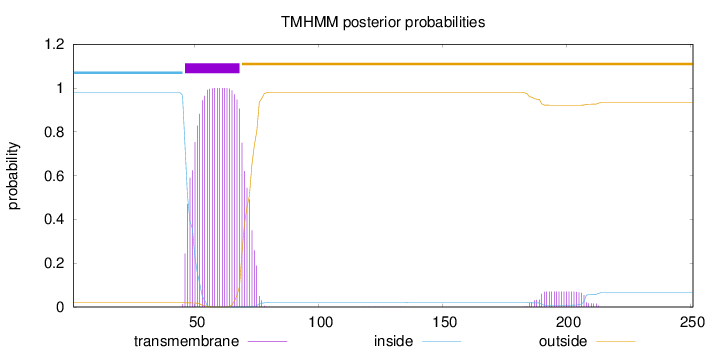
Topology
Length:
251
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
24.87405
Exp number, first 60 AAs:
12.29923
Total prob of N-in:
0.97992
POSSIBLE N-term signal
sequence
inside
1 - 45
TMhelix
46 - 68
outside
69 - 251
Population Genetic Test Statistics
Pi
312.182027
Theta
185.279462
Tajima's D
2.47108
CLR
0.148352
CSRT
0.943902804859757
Interpretation
Uncertain
