Gene
KWMTBOMO09705
Pre Gene Modal
BGIBMGA012888
Annotation
PREDICTED:_very-long-chain_3-oxoacyl-CoA_reductase_[Plutella_xylostella]
Full name
Very-long-chain 3-oxoacyl-CoA reductase
Alternative Name
17-beta-hydroxysteroid dehydrogenase 12
3-ketoacyl-CoA reductase
Estradiol 17-beta-dehydrogenase 12
3-ketoacyl-CoA reductase
Estradiol 17-beta-dehydrogenase 12
Location in the cell
PlasmaMembrane Reliability : 3.566
Sequence
CDS
ATGGCCCTATCGAATCTAGAAAAACTCGGAATATTATTTGTTGCATTTTTTATTTTTTATTGCCTTAAATTCTTGTGGCATTTCTTGTATACGTACGCCATTGGACCTGCAGTGCATTGTGTTGATTTCAAGTCCAAAGGGAAATGGGCACTTGTGACTGGATGCACTGATGGAATCGGAAAAGAATATGCTAAAGAGTTAGCCGCCCGTGGATGTGACATTGTTCTAGTCAGTCGATCTCTTGATAAACTAAAAGCAACAGCTGAAGAAATTGAGAAAGAATACAAGGTAGCCACGAAGATAATCCAAGCGGACTTTAGCGAGGACGATAAGATTTACGAGAACATTGAGAAGGAAATTGCTGGCCTGGAAATCGGTACTTTGGTCAATAATGTCGGAGTTTCCTATACGTATCCTGAATATTTCTTGGACCTGCCAGAGTGGGATAAGCTCATCCCAACTCTGATCAAAGCTAATGTGGTCGCTGTAACAAAGATGACACGCATCGTTTTACCCGAAATGGTGAAGAGAGAAAAGGGCGTTGTTATCAATATCGGGTCTGCATCTTCGATCATACCGAGTCCGTTGTTGACTGTCTACGCTGCTACTAAGGCATACGTCGATAAGTTCTCGGAGGGTCTCGATATGGAGTACTCGAAGAAGGGTATCGTTGTACAATGTATCCTACCCGGTTTCGTGTGCTCGAACATGTCCGGTATTCGTCGTAGCTCGCTATTCGCGCCCACGGCCAAGGTCTTCGTCCAATCGGCGATCAAGCTCGTCGGCACGACGACCAGGAACACTGGATACCTGCCCCACGCGGCCTTCGTGAACATCGTGAACAGCATCTACGAAACAGCAAACAGCTTCGGCACCTGGCTCGTCGTCAGATCCATGGAGAACAGCCGCAGGAAACTACTCAAAAAGACTGGAAAATTAGACGCTTAG
Protein
MALSNLEKLGILFVAFFIFYCLKFLWHFLYTYAIGPAVHCVDFKSKGKWALVTGCTDGIGKEYAKELAARGCDIVLVSRSLDKLKATAEEIEKEYKVATKIIQADFSEDDKIYENIEKEIAGLEIGTLVNNVGVSYTYPEYFLDLPEWDKLIPTLIKANVVAVTKMTRIVLPEMVKREKGVVINIGSASSIIPSPLLTVYAATKAYVDKFSEGLDMEYSKKGIVVQCILPGFVCSNMSGIRRSSLFAPTAKVFVQSAIKLVGTTTRNTGYLPHAAFVNIVNSIYETANSFGTWLVVRSMENSRRKLLKKTGKLDA
Summary
Description
Catalyzes the second of the four reactions of the long-chain fatty acids elongation cycle. This endoplasmic reticulum-bound enzymatic process, allows the addition of two carbons to the chain of long- and very long-chain fatty acids/VLCFAs per cycle. This enzyme has a 3-ketoacyl-CoA reductase activity, reducing 3-ketoacyl-CoA to 3-hydroxyacyl-CoA, within each cycle of fatty acid elongation. Thereby, it may participate in the production of VLCFAs of different chain lengths that are involved in multiple biological processes as precursors of membrane lipids and lipid mediators. May also catalyze the transformation of estrone (E1) into estradiol (E2) and play a role in estrogen formation.
Catalytic Activity
a very-long-chain (3R)-hydroxyacyl-CoA + NADP(+) = a very-long-chain 3-oxoacyl-CoA + H(+) + NADPH
17beta-estradiol + NAD(+) = estrone + H(+) + NADH
17beta-estradiol + NADP(+) = estrone + H(+) + NADPH
17beta-estradiol + NAD(+) = estrone + H(+) + NADH
17beta-estradiol + NADP(+) = estrone + H(+) + NADPH
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Belongs to the peptidase C1 family.
Belongs to the short-chain dehydrogenases/reductases (SDR) family. 17-beta-HSD 3 subfamily.
Belongs to the peptidase C1 family.
Belongs to the short-chain dehydrogenases/reductases (SDR) family. 17-beta-HSD 3 subfamily.
Keywords
Complete proteome
Endoplasmic reticulum
Lipid biosynthesis
Lipid metabolism
Membrane
NADP
Oxidoreductase
Reference proteome
Steroid biosynthesis
Transmembrane
Transmembrane helix
Feature
chain Very-long-chain 3-oxoacyl-CoA reductase
Uniprot
H9JTM6
A0A212FN25
S4NN93
A0A194PZV6
A0A2H1VMY3
A0A437BAX0
+ More
A0A1Q3G5H7 A0A194R1L7 A0A2A4K457 A0A067R7X3 D6W6K7 A0A1W4WFB5 E0VZ61 A0A2J7PXW4 V5IAV4 A0A1Y1MEY7 A0A182N9W6 A0A182QPS6 A0A182KZG1 A0A182XIF4 Q7PHB8 A0A182HYS4 U5ES67 A0A182JR85 A0A182Y8P6 A0A182RE10 A0A182W3J3 A0A084WTW3 A0A182IQ95 A0A182PHX0 T1I1N3 A0A226E5A4 A0A2M4ALT4 A0A1B6I742 A0A2M4ALL5 A0A2M4AMI8 E9IW58 W5JBX2 A0A2M3Z5K4 A0A0L0BZ82 A0A2M4BTK5 A0A2M3Z6D1 A0A224XUN0 A0A1S4ET94 A0A1B6M7J1 A0A1L8DYW5 A0A1L8DYZ8 A0A182M059 A0A0A9YXX0 A0A146KWV8 A0A1I8NH29 A0A0T6B503 B7PCL8 A0A401SB71 A0A131XVG3 B3MR02 B4NCV7 A0A336KD27 Q9W3N1 A0A0K8RJP6 A0A1W4VII7 B4L5D1 B3NTU8 B4Q0Q6 A0A1B6HDE9 B4MAC6 A0A0M4EIC5 A0A336LYP9 A0A1J1HPY6 B4R687 B4IL51 M3XF85 A0A1B6FVY4 A0A1B6IR56 A0A1B6HER9 A0A1I8PSS8 A0A151I4D2 A0A158N9V4 A0A218VA69 A0A224YS32 A0A131YZE9 A0A182F914 A0A023ENC8 A0A023EPQ8 A0A131XHF9 A0A2U4CDR4 Q6P7R8 A0A336LVR6 A0A2S2Q7I2 A0A340X3G0 W5NS44 F6Q4Z8 A0A1Z5LCX1 A0A3Q7W422 A0A2Y9PLY4 A0A2P8YXL4 A0A384CWR3 O57314 A0A195E1Q2 A0A151II76
A0A1Q3G5H7 A0A194R1L7 A0A2A4K457 A0A067R7X3 D6W6K7 A0A1W4WFB5 E0VZ61 A0A2J7PXW4 V5IAV4 A0A1Y1MEY7 A0A182N9W6 A0A182QPS6 A0A182KZG1 A0A182XIF4 Q7PHB8 A0A182HYS4 U5ES67 A0A182JR85 A0A182Y8P6 A0A182RE10 A0A182W3J3 A0A084WTW3 A0A182IQ95 A0A182PHX0 T1I1N3 A0A226E5A4 A0A2M4ALT4 A0A1B6I742 A0A2M4ALL5 A0A2M4AMI8 E9IW58 W5JBX2 A0A2M3Z5K4 A0A0L0BZ82 A0A2M4BTK5 A0A2M3Z6D1 A0A224XUN0 A0A1S4ET94 A0A1B6M7J1 A0A1L8DYW5 A0A1L8DYZ8 A0A182M059 A0A0A9YXX0 A0A146KWV8 A0A1I8NH29 A0A0T6B503 B7PCL8 A0A401SB71 A0A131XVG3 B3MR02 B4NCV7 A0A336KD27 Q9W3N1 A0A0K8RJP6 A0A1W4VII7 B4L5D1 B3NTU8 B4Q0Q6 A0A1B6HDE9 B4MAC6 A0A0M4EIC5 A0A336LYP9 A0A1J1HPY6 B4R687 B4IL51 M3XF85 A0A1B6FVY4 A0A1B6IR56 A0A1B6HER9 A0A1I8PSS8 A0A151I4D2 A0A158N9V4 A0A218VA69 A0A224YS32 A0A131YZE9 A0A182F914 A0A023ENC8 A0A023EPQ8 A0A131XHF9 A0A2U4CDR4 Q6P7R8 A0A336LVR6 A0A2S2Q7I2 A0A340X3G0 W5NS44 F6Q4Z8 A0A1Z5LCX1 A0A3Q7W422 A0A2Y9PLY4 A0A2P8YXL4 A0A384CWR3 O57314 A0A195E1Q2 A0A151II76
EC Number
1.1.1.330
Pubmed
19121390
22118469
23622113
26354079
24845553
18362917
+ More
19820115 20566863 28004739 20966253 12364791 14747013 17210077 25244985 24438588 21282665 20920257 23761445 26108605 25401762 26823975 25315136 30297745 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 17550304 17975172 21347285 28797301 26830274 24945155 26483478 28049606 15489334 20809919 19892987 28528879 29403074 24813606
19820115 20566863 28004739 20966253 12364791 14747013 17210077 25244985 24438588 21282665 20920257 23761445 26108605 25401762 26823975 25315136 30297745 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 17550304 17975172 21347285 28797301 26830274 24945155 26483478 28049606 15489334 20809919 19892987 28528879 29403074 24813606
EMBL
BABH01003520
BABH01003521
AGBW02007648
OWR55138.1
GAIX01014031
JAA78529.1
+ More
KQ459582 KPI98862.1 ODYU01003423 SOQ42167.1 RSAL01000100 RVE47541.1 GEYN01000182 JAV44807.1 KQ460878 KPJ11582.1 NWSH01000202 PCG78460.1 KK852819 KDR15624.1 KQ971307 EFA10853.1 DS235849 EEB18667.1 NEVH01020852 PNF21189.1 GALX01000530 JAB67936.1 GEZM01033612 JAV84213.1 AXCN02001202 AAAB01008898 EAA44603.5 APCN01005404 GANO01003389 JAB56482.1 ATLV01026924 KE525420 KFB53657.1 ACPB03005947 LNIX01000006 OXA52609.1 GGFK01008247 MBW41568.1 GECU01024981 JAS82725.1 GGFK01008177 MBW41498.1 GGFK01008507 MBW41828.1 GL766449 EFZ15287.1 ADMH02001667 ETN61491.1 GGFM01002977 MBW23728.1 JRES01001123 KNC25335.1 GGFJ01007191 MBW56332.1 GGFM01003332 MBW24083.1 GFTR01004613 JAW11813.1 GEBQ01008114 JAT31863.1 GFDF01002436 JAV11648.1 GFDF01002435 JAV11649.1 AXCM01000198 GBHO01009189 JAG34415.1 GDHC01017668 JAQ00961.1 LJIG01009859 KRT82238.1 ABJB010459395 ABJB010551348 ABJB010605370 ABJB010721098 ABJB010739574 DS684503 EEC04340.1 BEZZ01000172 GCC27639.1 GEFM01005484 JAP70312.1 CH902622 EDV34207.1 CH964239 EDW82666.1 UFQS01000234 UFQT01000234 SSX01762.1 SSX22140.1 AE014298 AY121708 AAF46291.1 AAM52035.1 GADI01002481 JAA71327.1 CH933811 EDW06390.1 CH954180 EDV47511.1 KQS30546.1 CM000162 EDX02327.1 GECU01034932 JAS72774.1 CH940655 EDW66185.1 KRF82589.1 KRF82591.1 CP012528 ALC48616.1 SSX01763.1 SSX22141.1 CVRI01000016 CRK90101.1 CM000366 EDX17364.1 CH480865 EDW53668.1 AANG04004363 GECZ01015403 JAS54366.1 GECU01018295 JAS89411.1 GECU01034528 JAS73178.1 KQ976461 KYM84702.1 ADTU01009655 MUZQ01000024 OWK62591.1 GFPF01005476 MAA16622.1 GEDV01004519 JAP84038.1 JXUM01001677 JXUM01001678 JXUM01001679 JXUM01001680 JXUM01001681 GAPW01002675 KQ560126 JAC10923.1 KXJ84333.1 GAPW01002603 JAC10995.1 GEFH01002048 JAP66533.1 U81186 BC061543 SSX01764.1 SSX22142.1 GGMS01003949 MBY73152.1 AMGL01033226 AMGL01033227 AMGL01033228 AMGL01033229 AMGL01033230 GFJQ02001912 JAW05058.1 PYGN01000299 PSN48986.1 AB009304 KQ979814 KYN19021.1 KQ977510 KYN02253.1
KQ459582 KPI98862.1 ODYU01003423 SOQ42167.1 RSAL01000100 RVE47541.1 GEYN01000182 JAV44807.1 KQ460878 KPJ11582.1 NWSH01000202 PCG78460.1 KK852819 KDR15624.1 KQ971307 EFA10853.1 DS235849 EEB18667.1 NEVH01020852 PNF21189.1 GALX01000530 JAB67936.1 GEZM01033612 JAV84213.1 AXCN02001202 AAAB01008898 EAA44603.5 APCN01005404 GANO01003389 JAB56482.1 ATLV01026924 KE525420 KFB53657.1 ACPB03005947 LNIX01000006 OXA52609.1 GGFK01008247 MBW41568.1 GECU01024981 JAS82725.1 GGFK01008177 MBW41498.1 GGFK01008507 MBW41828.1 GL766449 EFZ15287.1 ADMH02001667 ETN61491.1 GGFM01002977 MBW23728.1 JRES01001123 KNC25335.1 GGFJ01007191 MBW56332.1 GGFM01003332 MBW24083.1 GFTR01004613 JAW11813.1 GEBQ01008114 JAT31863.1 GFDF01002436 JAV11648.1 GFDF01002435 JAV11649.1 AXCM01000198 GBHO01009189 JAG34415.1 GDHC01017668 JAQ00961.1 LJIG01009859 KRT82238.1 ABJB010459395 ABJB010551348 ABJB010605370 ABJB010721098 ABJB010739574 DS684503 EEC04340.1 BEZZ01000172 GCC27639.1 GEFM01005484 JAP70312.1 CH902622 EDV34207.1 CH964239 EDW82666.1 UFQS01000234 UFQT01000234 SSX01762.1 SSX22140.1 AE014298 AY121708 AAF46291.1 AAM52035.1 GADI01002481 JAA71327.1 CH933811 EDW06390.1 CH954180 EDV47511.1 KQS30546.1 CM000162 EDX02327.1 GECU01034932 JAS72774.1 CH940655 EDW66185.1 KRF82589.1 KRF82591.1 CP012528 ALC48616.1 SSX01763.1 SSX22141.1 CVRI01000016 CRK90101.1 CM000366 EDX17364.1 CH480865 EDW53668.1 AANG04004363 GECZ01015403 JAS54366.1 GECU01018295 JAS89411.1 GECU01034528 JAS73178.1 KQ976461 KYM84702.1 ADTU01009655 MUZQ01000024 OWK62591.1 GFPF01005476 MAA16622.1 GEDV01004519 JAP84038.1 JXUM01001677 JXUM01001678 JXUM01001679 JXUM01001680 JXUM01001681 GAPW01002675 KQ560126 JAC10923.1 KXJ84333.1 GAPW01002603 JAC10995.1 GEFH01002048 JAP66533.1 U81186 BC061543 SSX01764.1 SSX22142.1 GGMS01003949 MBY73152.1 AMGL01033226 AMGL01033227 AMGL01033228 AMGL01033229 AMGL01033230 GFJQ02001912 JAW05058.1 PYGN01000299 PSN48986.1 AB009304 KQ979814 KYN19021.1 KQ977510 KYN02253.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000283053
UP000053240
UP000218220
+ More
UP000027135 UP000007266 UP000192223 UP000009046 UP000235965 UP000075884 UP000075886 UP000075882 UP000076407 UP000007062 UP000075840 UP000075881 UP000076408 UP000075900 UP000075920 UP000030765 UP000075880 UP000075885 UP000015103 UP000198287 UP000000673 UP000037069 UP000079169 UP000075883 UP000095301 UP000001555 UP000287033 UP000007801 UP000007798 UP000000803 UP000192221 UP000009192 UP000008711 UP000002282 UP000008792 UP000092553 UP000183832 UP000000304 UP000001292 UP000011712 UP000095300 UP000078540 UP000005205 UP000197619 UP000069272 UP000069940 UP000249989 UP000245320 UP000002494 UP000265300 UP000002356 UP000002281 UP000286642 UP000248483 UP000245037 UP000261680 UP000291021 UP000078492 UP000078542
UP000027135 UP000007266 UP000192223 UP000009046 UP000235965 UP000075884 UP000075886 UP000075882 UP000076407 UP000007062 UP000075840 UP000075881 UP000076408 UP000075900 UP000075920 UP000030765 UP000075880 UP000075885 UP000015103 UP000198287 UP000000673 UP000037069 UP000079169 UP000075883 UP000095301 UP000001555 UP000287033 UP000007801 UP000007798 UP000000803 UP000192221 UP000009192 UP000008711 UP000002282 UP000008792 UP000092553 UP000183832 UP000000304 UP000001292 UP000011712 UP000095300 UP000078540 UP000005205 UP000197619 UP000069272 UP000069940 UP000249989 UP000245320 UP000002494 UP000265300 UP000002356 UP000002281 UP000286642 UP000248483 UP000245037 UP000261680 UP000291021 UP000078492 UP000078542
Interpro
ProteinModelPortal
H9JTM6
A0A212FN25
S4NN93
A0A194PZV6
A0A2H1VMY3
A0A437BAX0
+ More
A0A1Q3G5H7 A0A194R1L7 A0A2A4K457 A0A067R7X3 D6W6K7 A0A1W4WFB5 E0VZ61 A0A2J7PXW4 V5IAV4 A0A1Y1MEY7 A0A182N9W6 A0A182QPS6 A0A182KZG1 A0A182XIF4 Q7PHB8 A0A182HYS4 U5ES67 A0A182JR85 A0A182Y8P6 A0A182RE10 A0A182W3J3 A0A084WTW3 A0A182IQ95 A0A182PHX0 T1I1N3 A0A226E5A4 A0A2M4ALT4 A0A1B6I742 A0A2M4ALL5 A0A2M4AMI8 E9IW58 W5JBX2 A0A2M3Z5K4 A0A0L0BZ82 A0A2M4BTK5 A0A2M3Z6D1 A0A224XUN0 A0A1S4ET94 A0A1B6M7J1 A0A1L8DYW5 A0A1L8DYZ8 A0A182M059 A0A0A9YXX0 A0A146KWV8 A0A1I8NH29 A0A0T6B503 B7PCL8 A0A401SB71 A0A131XVG3 B3MR02 B4NCV7 A0A336KD27 Q9W3N1 A0A0K8RJP6 A0A1W4VII7 B4L5D1 B3NTU8 B4Q0Q6 A0A1B6HDE9 B4MAC6 A0A0M4EIC5 A0A336LYP9 A0A1J1HPY6 B4R687 B4IL51 M3XF85 A0A1B6FVY4 A0A1B6IR56 A0A1B6HER9 A0A1I8PSS8 A0A151I4D2 A0A158N9V4 A0A218VA69 A0A224YS32 A0A131YZE9 A0A182F914 A0A023ENC8 A0A023EPQ8 A0A131XHF9 A0A2U4CDR4 Q6P7R8 A0A336LVR6 A0A2S2Q7I2 A0A340X3G0 W5NS44 F6Q4Z8 A0A1Z5LCX1 A0A3Q7W422 A0A2Y9PLY4 A0A2P8YXL4 A0A384CWR3 O57314 A0A195E1Q2 A0A151II76
A0A1Q3G5H7 A0A194R1L7 A0A2A4K457 A0A067R7X3 D6W6K7 A0A1W4WFB5 E0VZ61 A0A2J7PXW4 V5IAV4 A0A1Y1MEY7 A0A182N9W6 A0A182QPS6 A0A182KZG1 A0A182XIF4 Q7PHB8 A0A182HYS4 U5ES67 A0A182JR85 A0A182Y8P6 A0A182RE10 A0A182W3J3 A0A084WTW3 A0A182IQ95 A0A182PHX0 T1I1N3 A0A226E5A4 A0A2M4ALT4 A0A1B6I742 A0A2M4ALL5 A0A2M4AMI8 E9IW58 W5JBX2 A0A2M3Z5K4 A0A0L0BZ82 A0A2M4BTK5 A0A2M3Z6D1 A0A224XUN0 A0A1S4ET94 A0A1B6M7J1 A0A1L8DYW5 A0A1L8DYZ8 A0A182M059 A0A0A9YXX0 A0A146KWV8 A0A1I8NH29 A0A0T6B503 B7PCL8 A0A401SB71 A0A131XVG3 B3MR02 B4NCV7 A0A336KD27 Q9W3N1 A0A0K8RJP6 A0A1W4VII7 B4L5D1 B3NTU8 B4Q0Q6 A0A1B6HDE9 B4MAC6 A0A0M4EIC5 A0A336LYP9 A0A1J1HPY6 B4R687 B4IL51 M3XF85 A0A1B6FVY4 A0A1B6IR56 A0A1B6HER9 A0A1I8PSS8 A0A151I4D2 A0A158N9V4 A0A218VA69 A0A224YS32 A0A131YZE9 A0A182F914 A0A023ENC8 A0A023EPQ8 A0A131XHF9 A0A2U4CDR4 Q6P7R8 A0A336LVR6 A0A2S2Q7I2 A0A340X3G0 W5NS44 F6Q4Z8 A0A1Z5LCX1 A0A3Q7W422 A0A2Y9PLY4 A0A2P8YXL4 A0A384CWR3 O57314 A0A195E1Q2 A0A151II76
PDB
4BMV
E-value=8.59185e-23,
Score=263
Ontologies
PATHWAY
GO
Topology
Subcellular location
Endoplasmic reticulum membrane
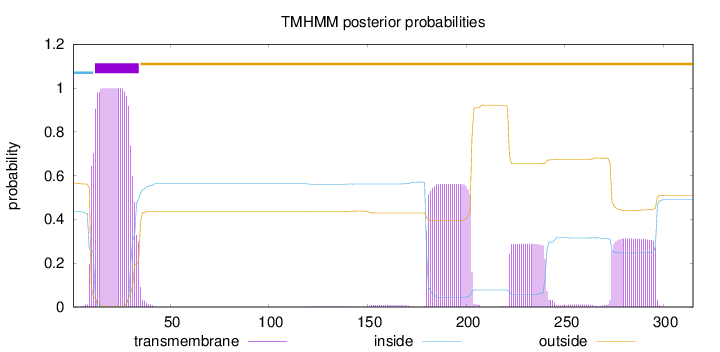
Length:
315
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
47.72605
Exp number, first 60 AAs:
21.91992
Total prob of N-in:
0.43610
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 34
outside
35 - 315
Population Genetic Test Statistics
Pi
288.117633
Theta
177.166712
Tajima's D
1.839293
CLR
0.167713
CSRT
0.856907154642268
Interpretation
Uncertain
