Gene
KWMTBOMO09695
Pre Gene Modal
BGIBMGA012882
Annotation
PREDICTED:_TWiK_family_of_potassium_channels_protein_9-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.579
Sequence
CDS
ATGGATATAGATCATGTGGTGCTGGATAAAGAAGCAATCAATCGTTTCGAACAGTATACGTTGGCAAAGCCATCGCTAAACGGGGGGATCCCTTTGGACAGATGCCAAGACGCCGACTCCATGCTCTCTAATAGCAGGAAGAGAGCACAAATATCACGCCAAAAAGTTGTTTGCTGCTTTTGTTTCAAAACTACGAAATACAGGAGAAAACAATTCATTATAGGATCGCTTACAAATGTCGTTATATTCGCTATTTTGTTGGCGTATACTTTCCTCGGCTCTTTTGTATTCTTAGCTATAGAGGGAGGTGTGGAAATACCGGTCAGACCTCGCCTGTTACCGGACAGACAAAAAATAAACAACAAAAGCGAGAATGCCACAAGAAAGAAATACCCGGACGACGATTACGACTACGGTAACTTGACATTATCAGCGAATTATTTCTGGGACGCGAGAAGCAGAGCAGTCGAAAACATTTGGGAGATAACCGTCTCACTAAATATATTGTACAGAGAGAATTGGACGCGGTTGGCTGCACAGGAGATACTGAAATTTCAAAACGAGCTGGTGCAGAGGGTGACTACTGAAATGGCGTCACAATATGGCGTTAGTTACAGAGAAATGGCTCTTGGAGAATTTGGCGGGAACGAAGCCAGTAACCATTATGAGGAGCATGAGTGGAATTTAGCCTTAGCATTTTTCTACTCGCTCACTGTTTTGACTACTATAGGCTATGGAAATATTGCTCCACGAACGATACTTGGCAAAGGAGTTACAATTTTATACGCTTTAATCGGGATACCACTAACATTGGTCTACTTATCCAGTGTGGGATCTCTACTATCTAAAATGGCTAGAAGTGTCTTCAGTAAAGCCCTTTGCTGTTGCCTTTGTTCTAAATGTGGCTACTGCTGTTATGATGAGAGGAGAATGGCGGAGAAGGAAAGACGCATGAAACTTAAGAGGCAGCAAGAGGAGATGCTCAACAGTCAAAACACCAGCAAAACGCCCACTGAAGAGTGCTACGTTTTAAAGCCAGACAGTCAGAAAGACTCGTCTCTATCAGAAAGGCCGACGACGAGCACTGCCAAAGATGATATCATAAGTTGGCCTGACACAGATTCGAAGCTATCTATGCACGGTTTAAGTATATTAGCCCCAGTTTTCCTGTGTCTCGTCGCCATATTCATTTACATAGTCGCAGGGGCCGTTGTTCTTTTTAAATTGGACGATTTAAGTATTATCGACGGTTTTTATTTTTGTTTCATGGCATTAAGTACGATTGGCTTTGGTAACATCGTTCCCGGGATGAGCTACACGACAATCCACGGAGTTAGATACTATACGATTAATTCGACAACGCTTTGGTTTTGTTCGGCCTATACACTGACCGGTCTCGCTTTGACCGCGATGTGTTTCGGTGTTATCCACGACGAGATAATATATAGAATTAAGCACCAGCAAAAGGAATGGTCTCAGAAGGGAAACACTGTTAATGATGAGAGCAAATCGGAGAACTTGTACAGTATAATGGAGGATAGCTCCGTAGAACGTAGTAGAATCGAAATCGACAATAGGGAGGTAGTTATTAGTGTAGATGATGTGTCAGGAGCAATGGATTCGAGTGCCGAGTTCCCCCCACCACCCCCCGAAGAAGAAATTATAGCTATTGTAACGAAAAAAGAACCTAAAGGTTTCGGCAATATGGTTGCATATAACGCTCTACCAGAAATGCTCGAGCATGTGAACACGTAA
Protein
MDIDHVVLDKEAINRFEQYTLAKPSLNGGIPLDRCQDADSMLSNSRKRAQISRQKVVCCFCFKTTKYRRKQFIIGSLTNVVIFAILLAYTFLGSFVFLAIEGGVEIPVRPRLLPDRQKINNKSENATRKKYPDDDYDYGNLTLSANYFWDARSRAVENIWEITVSLNILYRENWTRLAAQEILKFQNELVQRVTTEMASQYGVSYREMALGEFGGNEASNHYEEHEWNLALAFFYSLTVLTTIGYGNIAPRTILGKGVTILYALIGIPLTLVYLSSVGSLLSKMARSVFSKALCCCLCSKCGYCCYDERRMAEKERRMKLKRQQEEMLNSQNTSKTPTEECYVLKPDSQKDSSLSERPTTSTAKDDIISWPDTDSKLSMHGLSILAPVFLCLVAIFIYIVAGAVVLFKLDDLSIIDGFYFCFMALSTIGFGNIVPGMSYTTIHGVRYYTINSTTLWFCSAYTLTGLALTAMCFGVIHDEIIYRIKHQQKEWSQKGNTVNDESKSENLYSIMEDSSVERSRIEIDNREVVISVDDVSGAMDSSAEFPPPPPEEEIIAIVTKKEPKGFGNMVAYNALPEMLEHVNT
Summary
Similarity
Belongs to the two pore domain potassium channel (TC 1.A.1.8) family.
Uniprot
H9JTM0
A0A3S2LIQ5
A0A0L7L7U6
A0A2H1X3T4
A0A2A4JM27
A0A2W1BAR8
+ More
A0A194Q1I5 A0A194R1M7 A0A212FN32 A0A194R6U4 H9JTT7 A0A0L7KMZ7 A0A3S2NY61 A0A194Q0N1 A0A212FN26 A0A2W1BHB4 A0A2H1VRM0 D2A6I4 A0A1J1ID08 A0A182MLI1 A0A182W079 A0A182N624 A0A182RNP6 A0A1S4HDT8 A0A182LFS8 A0A182Y4C5 A0A182PAA6 W5JF99 A0A3L8DE05 A0A182F7F2 A0A154PFT0 A0A182U5S3 A0A158NJU8 A0A182K1R4 B4M3H8 B4L462 A0A3B0K933 A0A2H8TI30 A0A2S2NWI9 J9JQB1 A0A2R7WRI0 A0A182GEM1 A0A182FLS6
A0A194Q1I5 A0A194R1M7 A0A212FN32 A0A194R6U4 H9JTT7 A0A0L7KMZ7 A0A3S2NY61 A0A194Q0N1 A0A212FN26 A0A2W1BHB4 A0A2H1VRM0 D2A6I4 A0A1J1ID08 A0A182MLI1 A0A182W079 A0A182N624 A0A182RNP6 A0A1S4HDT8 A0A182LFS8 A0A182Y4C5 A0A182PAA6 W5JF99 A0A3L8DE05 A0A182F7F2 A0A154PFT0 A0A182U5S3 A0A158NJU8 A0A182K1R4 B4M3H8 B4L462 A0A3B0K933 A0A2H8TI30 A0A2S2NWI9 J9JQB1 A0A2R7WRI0 A0A182GEM1 A0A182FLS6
Pubmed
EMBL
BABH01003557
RSAL01000100
RVE47532.1
JTDY01002366
KOB71567.1
ODYU01013224
+ More
SOQ59918.1 NWSH01001064 PCG72836.1 KZ150267 PZC71701.1 KQ459582 KPI98854.1 KQ460878 KPJ11592.1 AGBW02007648 OWR55145.1 KPJ11581.1 BABH01003515 JTDY01008277 KOB64648.1 RVE47542.1 KPI98863.1 OWR55137.1 KZ150128 PZC73084.1 ODYU01004008 SOQ43447.1 KQ971348 EFA04915.1 CVRI01000047 CRK98167.1 AXCM01001083 AAAB01008859 ADMH02001371 ETN62761.1 QOIP01000009 RLU18665.1 KQ434893 KZC10672.1 ADTU01001990 ADTU01001991 ADTU01001992 ADTU01001993 ADTU01001994 ADTU01001995 CH940651 EDW65353.2 CH933810 EDW07340.2 OUUW01000036 SPP89863.1 GFXV01001982 MBW13787.1 GGMR01008467 MBY21086.1 ABLF02035901 ABLF02035904 ABLF02035907 ABLF02035909 ABLF02035910 KK855358 PTY22164.1 JXUM01058168 JXUM01058169 JXUM01058170 JXUM01058171 JXUM01058172 JXUM01058173 KQ561990 KXJ76972.1
SOQ59918.1 NWSH01001064 PCG72836.1 KZ150267 PZC71701.1 KQ459582 KPI98854.1 KQ460878 KPJ11592.1 AGBW02007648 OWR55145.1 KPJ11581.1 BABH01003515 JTDY01008277 KOB64648.1 RVE47542.1 KPI98863.1 OWR55137.1 KZ150128 PZC73084.1 ODYU01004008 SOQ43447.1 KQ971348 EFA04915.1 CVRI01000047 CRK98167.1 AXCM01001083 AAAB01008859 ADMH02001371 ETN62761.1 QOIP01000009 RLU18665.1 KQ434893 KZC10672.1 ADTU01001990 ADTU01001991 ADTU01001992 ADTU01001993 ADTU01001994 ADTU01001995 CH940651 EDW65353.2 CH933810 EDW07340.2 OUUW01000036 SPP89863.1 GFXV01001982 MBW13787.1 GGMR01008467 MBY21086.1 ABLF02035901 ABLF02035904 ABLF02035907 ABLF02035909 ABLF02035910 KK855358 PTY22164.1 JXUM01058168 JXUM01058169 JXUM01058170 JXUM01058171 JXUM01058172 JXUM01058173 KQ561990 KXJ76972.1
Proteomes
UP000005204
UP000283053
UP000037510
UP000218220
UP000053268
UP000053240
+ More
UP000007151 UP000007266 UP000183832 UP000075883 UP000075920 UP000075884 UP000075900 UP000075882 UP000076408 UP000075885 UP000000673 UP000279307 UP000069272 UP000076502 UP000075902 UP000005205 UP000075881 UP000008792 UP000009192 UP000268350 UP000007819 UP000069940 UP000249989
UP000007151 UP000007266 UP000183832 UP000075883 UP000075920 UP000075884 UP000075900 UP000075882 UP000076408 UP000075885 UP000000673 UP000279307 UP000069272 UP000076502 UP000075902 UP000005205 UP000075881 UP000008792 UP000009192 UP000268350 UP000007819 UP000069940 UP000249989
Pfam
PF07885 Ion_trans_2
ProteinModelPortal
H9JTM0
A0A3S2LIQ5
A0A0L7L7U6
A0A2H1X3T4
A0A2A4JM27
A0A2W1BAR8
+ More
A0A194Q1I5 A0A194R1M7 A0A212FN32 A0A194R6U4 H9JTT7 A0A0L7KMZ7 A0A3S2NY61 A0A194Q0N1 A0A212FN26 A0A2W1BHB4 A0A2H1VRM0 D2A6I4 A0A1J1ID08 A0A182MLI1 A0A182W079 A0A182N624 A0A182RNP6 A0A1S4HDT8 A0A182LFS8 A0A182Y4C5 A0A182PAA6 W5JF99 A0A3L8DE05 A0A182F7F2 A0A154PFT0 A0A182U5S3 A0A158NJU8 A0A182K1R4 B4M3H8 B4L462 A0A3B0K933 A0A2H8TI30 A0A2S2NWI9 J9JQB1 A0A2R7WRI0 A0A182GEM1 A0A182FLS6
A0A194Q1I5 A0A194R1M7 A0A212FN32 A0A194R6U4 H9JTT7 A0A0L7KMZ7 A0A3S2NY61 A0A194Q0N1 A0A212FN26 A0A2W1BHB4 A0A2H1VRM0 D2A6I4 A0A1J1ID08 A0A182MLI1 A0A182W079 A0A182N624 A0A182RNP6 A0A1S4HDT8 A0A182LFS8 A0A182Y4C5 A0A182PAA6 W5JF99 A0A3L8DE05 A0A182F7F2 A0A154PFT0 A0A182U5S3 A0A158NJU8 A0A182K1R4 B4M3H8 B4L462 A0A3B0K933 A0A2H8TI30 A0A2S2NWI9 J9JQB1 A0A2R7WRI0 A0A182GEM1 A0A182FLS6
PDB
4XDL
E-value=1.03779e-08,
Score=145
Ontologies
GO
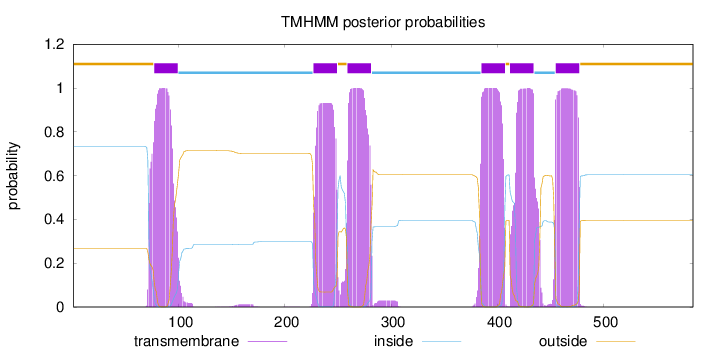
Topology
Length:
584
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
134.65872
Exp number, first 60 AAs:
0.00028
Total prob of N-in:
0.73254
outside
1 - 76
TMhelix
77 - 99
inside
100 - 226
TMhelix
227 - 249
outside
250 - 258
TMhelix
259 - 281
inside
282 - 384
TMhelix
385 - 407
outside
408 - 411
TMhelix
412 - 434
inside
435 - 454
TMhelix
455 - 477
outside
478 - 584
Population Genetic Test Statistics
Pi
265.708581
Theta
180.716881
Tajima's D
1.958153
CLR
0.198356
CSRT
0.874106294685266
Interpretation
Uncertain
