Gene
KWMTBOMO09692
Pre Gene Modal
BGIBMGA012880
Annotation
PREDICTED:_transcription_initiation_protein_SPT3_homolog_[Amyelois_transitella]
Full name
Transcription initiation protein SPT3 homolog
Alternative Name
SPT3-like protein
Location in the cell
Nuclear Reliability : 2.256
Sequence
CDS
ATGACTTACTATACTATGGAAACATGTGGAGAAGGGACGAACTTTCAAAAAGAAATATCGAACATGATGCACGGATTTGGAGATAACCCGAACCCTAACGCTGCAACGGTACTTTTAGTAGAAAATATTGTTTTGCAACAGCTTAGGAGTATCCTACAAGAAGGAATATTTAATTCTAACTTGAGAGGAAGTAAAACAATCACTAACTACGACATAATATACACAATGCGTAACAATACTGTAAAATTAAAACGTTTGCACGATTACCAGCAAAAACTGGATCAAATCGACAACAAATCACAAGTAAACGTCACAACATCAGCCGAAACTATATACTCTAATATTATACTAGAAGACGAGAAGGATCCGACAATGAAAAAAAAGAGAACACATATTGATGTGATAAAACTAATAGACGAAGCTGATGAAGTGAGTCAGATAAAATTCGATGCGATCGATTACTTAAGGAAAGTGCGAGCTTCGAAATTAACAGAGTCAATGTCTTTTGAAAAGTATGAAGCCTATCATAAAGCTAGATGTAGTTCTTTCCGTTGCAATGGCTATGGAATCGGTAAAGGCTTTATAAAATTAGAAAGGTGGATCAATCCAACCAAGGAACTCAAAATTAGTCACTCGGCATTAGAAGTATTGTGTTTTATTGCTTACGAAACTGTGGCTGAAATTGTTGATGCTGTGTATTTGATTCGACAAGATGCCAAAAGGAAACACGGAGACCCTTTTAGTAAATATGAAGGTGGATATTTCTGTAATCCCTTAAGCTTAAGTAATGCTGTTTATATAAAGTCAGGTTATGAAGGAGTGCCTGCAATAACAGTGGCTGAAGTGAGAGAGGCATTGCGTCGTTACTTTTGTCCTATAAATGGAATGAATGGATTATTTTATAGAAATATGCACATGGACTTACCAGCAAGATTTATTGCTTTATAA
Protein
MTYYTMETCGEGTNFQKEISNMMHGFGDNPNPNAATVLLVENIVLQQLRSILQEGIFNSNLRGSKTITNYDIIYTMRNNTVKLKRLHDYQQKLDQIDNKSQVNVTTSAETIYSNIILEDEKDPTMKKKRTHIDVIKLIDEADEVSQIKFDAIDYLRKVRASKLTESMSFEKYEAYHKARCSSFRCNGYGIGKGFIKLERWINPTKELKISHSALEVLCFIAYETVAEIVDAVYLIRQDAKRKHGDPFSKYEGGYFCNPLSLSNAVYIKSGYEGVPAITVAEVREALRRYFCPINGMNGLFYRNMHMDLPARFIAL
Summary
Description
Probable transcriptional activator.
Subunit
Component of the PCAF complex, at least composed of TADA2L/ADA2, SUPT3H, TADA3L/ADA3, TAF5L/PAF65-beta, TAF6L/PAF65-alpha, TAF10/TAFII30, TAF12/TAFII20, TAF9/TAFII31 and TRRAP. Associates with TAFII31 and GCN5L2. Component of the TFTC-HAT complex, at least composed of TAF5L, TAF6L, TADA3L, SUPT3H/SPT3, TAF2/TAFII150, TAF4/TAFII135, TAF5/TAFII100, GCN5L2/GCN5, TAF10 and TRRAP. Component of the STAGA transcription coactivator-HAT complex, at least composed of SUPT3H, GCN5L2, TAF5L, TAF6L, SUPT7L, TADA3L, TAD1L, TAF10, TAF12, TRRAP and TAF9. The STAGA core complex is associated with a subcomplex required for histone deubiquitination composed of ATXN7L3, ENY2 and USP22.
Similarity
Belongs to the SPT3 family.
Keywords
Acetylation
Activator
Alternative splicing
Complete proteome
Direct protein sequencing
Isopeptide bond
Nucleus
Polymorphism
Reference proteome
Transcription
Transcription regulation
Ubl conjugation
Feature
chain Transcription initiation protein SPT3 homolog
splice variant In isoform 2.
sequence variant In dbSNP:rs16872923.
splice variant In isoform 2.
sequence variant In dbSNP:rs16872923.
Uniprot
H9JTL8
A0A1E1WSF9
A0A194R1C3
A0A2A4JD18
S4PY51
A0A2H1VIX2
+ More
A0A212FN00 A0A0L7L2A4 A0A0L7LG43 A0A0L7L263 A0A1W4WUS4 D6WHX1 A0A0N0BKJ7 K7IP16 A0A2P8XS51 A0A232EUT9 A0A026WUS6 A0A293MT60 A0A2R5LD81 A0A0C9QSG8 A0A067RI08 A0A0L7QMR3 A0A154PF75 L7LQW8 E2A273 A0A1Z5L1F1 E2BNL7 A0A0J7K4Y7 G3MI98 A0A2K5ERL1 A0A224Z8T2 A0A2K6KKH5 A0A2K6NBV1 A0A2K5I8S4 A0A151IVP8 G1T7P0 E9IWP1 K7DEW9 A0A2J8Y4X1 G3RFB2 A0A024RD67 O75486 A0A2I3HJM7 A0A2K5ZLH1 A0A2K6V6B1 A0A2K6BFJ1 A0A158NN48 A0A2K5LG50 A0A2K5UI13 A0A2I3LH29 H9G0V2 A0A195FGS8 A0A131YW77 A0A182YJC1 Q5U608 U3D6N9 A0A023GML6 F4X369 A0A2R9AIQ8 A0A340XGK0 M3WBP7 A0A182WEJ1 A0A195BET6 A0A1E1X3Z4 A0A3B1KG57 A0A0P4W7L0 U5EGB3 N6T5T7 T1KYN1 A0A1S3FDC8 A0A2K6EMP3 A0A0P6J671 A0A182M2Q8 A0A182R6Z9 A0A084VID8 A0A2D4JV86 G3UR77 A0A3B3T3S8 A0A182PC97 A0A2D4I4Z9 A0A182J891 Q5ZIW0 A0A3Q2UIL6 A0A2A3EEV6 A0A2M4AY25 G5BNF1 A0A2R8MX91 A0A2U4BIY3 I3MPQ3 A0A151IKZ8 W5JVU5 A0A2Y9JIB1 A0A452J2P7 A0A182XKL5 A0A182VDM6 A0A182L4B0 A0A182TN70 A0A182HGX0
A0A212FN00 A0A0L7L2A4 A0A0L7LG43 A0A0L7L263 A0A1W4WUS4 D6WHX1 A0A0N0BKJ7 K7IP16 A0A2P8XS51 A0A232EUT9 A0A026WUS6 A0A293MT60 A0A2R5LD81 A0A0C9QSG8 A0A067RI08 A0A0L7QMR3 A0A154PF75 L7LQW8 E2A273 A0A1Z5L1F1 E2BNL7 A0A0J7K4Y7 G3MI98 A0A2K5ERL1 A0A224Z8T2 A0A2K6KKH5 A0A2K6NBV1 A0A2K5I8S4 A0A151IVP8 G1T7P0 E9IWP1 K7DEW9 A0A2J8Y4X1 G3RFB2 A0A024RD67 O75486 A0A2I3HJM7 A0A2K5ZLH1 A0A2K6V6B1 A0A2K6BFJ1 A0A158NN48 A0A2K5LG50 A0A2K5UI13 A0A2I3LH29 H9G0V2 A0A195FGS8 A0A131YW77 A0A182YJC1 Q5U608 U3D6N9 A0A023GML6 F4X369 A0A2R9AIQ8 A0A340XGK0 M3WBP7 A0A182WEJ1 A0A195BET6 A0A1E1X3Z4 A0A3B1KG57 A0A0P4W7L0 U5EGB3 N6T5T7 T1KYN1 A0A1S3FDC8 A0A2K6EMP3 A0A0P6J671 A0A182M2Q8 A0A182R6Z9 A0A084VID8 A0A2D4JV86 G3UR77 A0A3B3T3S8 A0A182PC97 A0A2D4I4Z9 A0A182J891 Q5ZIW0 A0A3Q2UIL6 A0A2A3EEV6 A0A2M4AY25 G5BNF1 A0A2R8MX91 A0A2U4BIY3 I3MPQ3 A0A151IKZ8 W5JVU5 A0A2Y9JIB1 A0A452J2P7 A0A182XKL5 A0A182VDM6 A0A182L4B0 A0A182TN70 A0A182HGX0
Pubmed
19121390
26354079
23622113
22118469
26227816
18362917
+ More
19820115 20075255 29403074 28648823 24508170 30249741 24845553 25576852 20798317 28528879 22216098 28797301 25362486 21993624 21282665 16136131 22398555 11181995 9787080 9674425 9726987 14702039 14574404 15489334 9885574 10373431 11564863 18206972 22814378 25218447 28112733 21347285 17431167 25319552 26830274 25244985 25243066 21719571 22722832 17975172 28503490 25329095 23537049 24438588 20838655 29240929 15592404 15642098 21993625 20920257 23761445 28562605 20966253
19820115 20075255 29403074 28648823 24508170 30249741 24845553 25576852 20798317 28528879 22216098 28797301 25362486 21993624 21282665 16136131 22398555 11181995 9787080 9674425 9726987 14702039 14574404 15489334 9885574 10373431 11564863 18206972 22814378 25218447 28112733 21347285 17431167 25319552 26830274 25244985 25243066 21719571 22722832 17975172 28503490 25329095 23537049 24438588 20838655 29240929 15592404 15642098 21993625 20920257 23761445 28562605 20966253
EMBL
BABH01003606
GDQN01001089
JAT89965.1
KQ460878
KPJ11593.1
NWSH01002011
+ More
PCG69444.1 GAIX01003898 JAA88662.1 ODYU01002820 SOQ40777.1 AGBW02007649 OWR55122.1 JTDY01003417 KOB69587.1 JTDY01001211 KOB74523.1 KOB69588.1 KQ971321 EFA00044.2 KQ435698 KOX80692.1 PYGN01001454 PSN34744.1 NNAY01002091 OXU22103.1 KK107106 QOIP01000002 EZA59416.1 RLU26132.1 GFWV01019264 MAA43992.1 GGLE01003181 MBY07307.1 GBYB01006679 JAG76446.1 KK852669 KDR18882.1 KQ414885 KOC59885.1 KQ434892 KZC10536.1 GACK01010618 JAA54416.1 GL435922 EFN72478.1 GFJQ02005764 JAW01206.1 GL449414 EFN82796.1 LBMM01014287 KMQ85246.1 JO841599 AEO33216.1 GFPF01012167 MAA23313.1 KQ980895 KYN11751.1 AAGW02018051 AAGW02018052 AAGW02018053 AAGW02018054 AAGW02018055 AAGW02018056 AAGW02018057 AAGW02018058 AAGW02018059 AAGW02018060 GL766597 EFZ14990.1 AACZ04022891 AACZ04022892 AACZ04022893 AACZ04022894 AACZ04022895 AACZ04022896 AACZ04022897 GABC01002300 GABF01010021 GABD01010751 GABE01005295 NBAG03000221 JAA09038.1 JAA12124.1 JAA22349.1 JAA39444.1 PNI77556.1 PNI77558.1 ABGA01248159 ABGA01248160 ABGA01248161 ABGA01248162 ABGA01248163 ABGA01248164 ABGA01248165 ABGA01248166 ABGA01248167 ABGA01248168 NDHI03003283 PNJ89325.1 PNJ89327.1 CABD030044832 CABD030044833 CABD030044834 CABD030044835 CABD030044836 CABD030044837 CABD030044838 CABD030044839 CABD030044840 CABD030044841 CH471081 EAX04276.1 AF064804 AF069734 AF073930 AK313874 AL096865 AL138880 AL161905 AL360272 AL499610 BC050384 ADFV01032095 ADFV01032096 ADFV01032097 ADFV01032098 ADFV01032099 ADFV01032100 ADFV01032101 ADFV01032102 ADFV01032103 ADFV01032104 ADTU01000299 ADTU01000300 AQIA01051756 AQIA01051757 AQIA01051758 AQIA01051759 AQIA01051760 AQIA01051761 AQIA01051762 AHZZ02024703 AHZZ02024704 AHZZ02024705 AHZZ02024706 AHZZ02024707 JSUE03030958 JSUE03030959 JSUE03030960 JSUE03030961 JSUE03030962 JSUE03030963 JSUE03030964 JU336758 JU477522 JV048467 AFE80511.1 AFH34326.1 AFI38538.1 KQ981606 KYN39581.1 GEDV01005350 JAP83207.1 BC035816 AAH35816.1 GAMS01002775 GAMR01008008 GAMP01010164 JAB20361.1 JAB25924.1 JAB42591.1 GBBM01001298 JAC34120.1 GL888609 EGI59050.1 AJFE02119515 AJFE02119516 AJFE02119517 AJFE02119518 AJFE02119519 AJFE02119520 AJFE02119521 AJFE02119522 AJFE02119523 AJFE02119524 AANG04003966 KQ976509 KYM82680.1 GFAC01005203 JAT93985.1 GDRN01076373 JAI62903.1 GANO01003514 JAB56357.1 APGK01043031 KB741011 KB631636 KI209682 ENN75554.1 ERL84883.1 ERL96074.1 CAEY01000714 GEBF01003892 JAN99740.1 AXCM01001816 ATLV01013353 KE524854 KFB37732.1 IACL01015259 IACL01015260 LAB00378.1 IACK01081177 IACK01081181 LAA79303.1 AADN05001052 AJ720674 CAG32333.1 KZ288266 PBC30267.1 GGFK01012372 MBW45693.1 JH171133 EHB10892.1 AGTP01010763 AGTP01010764 AGTP01010765 AGTP01010766 AGTP01010767 AGTP01010768 AGTP01010769 AGTP01010770 AGTP01010771 AGTP01010772 KQ977151 KYN05286.1 ADMH02000317 ETN67004.1 APCN01002367
PCG69444.1 GAIX01003898 JAA88662.1 ODYU01002820 SOQ40777.1 AGBW02007649 OWR55122.1 JTDY01003417 KOB69587.1 JTDY01001211 KOB74523.1 KOB69588.1 KQ971321 EFA00044.2 KQ435698 KOX80692.1 PYGN01001454 PSN34744.1 NNAY01002091 OXU22103.1 KK107106 QOIP01000002 EZA59416.1 RLU26132.1 GFWV01019264 MAA43992.1 GGLE01003181 MBY07307.1 GBYB01006679 JAG76446.1 KK852669 KDR18882.1 KQ414885 KOC59885.1 KQ434892 KZC10536.1 GACK01010618 JAA54416.1 GL435922 EFN72478.1 GFJQ02005764 JAW01206.1 GL449414 EFN82796.1 LBMM01014287 KMQ85246.1 JO841599 AEO33216.1 GFPF01012167 MAA23313.1 KQ980895 KYN11751.1 AAGW02018051 AAGW02018052 AAGW02018053 AAGW02018054 AAGW02018055 AAGW02018056 AAGW02018057 AAGW02018058 AAGW02018059 AAGW02018060 GL766597 EFZ14990.1 AACZ04022891 AACZ04022892 AACZ04022893 AACZ04022894 AACZ04022895 AACZ04022896 AACZ04022897 GABC01002300 GABF01010021 GABD01010751 GABE01005295 NBAG03000221 JAA09038.1 JAA12124.1 JAA22349.1 JAA39444.1 PNI77556.1 PNI77558.1 ABGA01248159 ABGA01248160 ABGA01248161 ABGA01248162 ABGA01248163 ABGA01248164 ABGA01248165 ABGA01248166 ABGA01248167 ABGA01248168 NDHI03003283 PNJ89325.1 PNJ89327.1 CABD030044832 CABD030044833 CABD030044834 CABD030044835 CABD030044836 CABD030044837 CABD030044838 CABD030044839 CABD030044840 CABD030044841 CH471081 EAX04276.1 AF064804 AF069734 AF073930 AK313874 AL096865 AL138880 AL161905 AL360272 AL499610 BC050384 ADFV01032095 ADFV01032096 ADFV01032097 ADFV01032098 ADFV01032099 ADFV01032100 ADFV01032101 ADFV01032102 ADFV01032103 ADFV01032104 ADTU01000299 ADTU01000300 AQIA01051756 AQIA01051757 AQIA01051758 AQIA01051759 AQIA01051760 AQIA01051761 AQIA01051762 AHZZ02024703 AHZZ02024704 AHZZ02024705 AHZZ02024706 AHZZ02024707 JSUE03030958 JSUE03030959 JSUE03030960 JSUE03030961 JSUE03030962 JSUE03030963 JSUE03030964 JU336758 JU477522 JV048467 AFE80511.1 AFH34326.1 AFI38538.1 KQ981606 KYN39581.1 GEDV01005350 JAP83207.1 BC035816 AAH35816.1 GAMS01002775 GAMR01008008 GAMP01010164 JAB20361.1 JAB25924.1 JAB42591.1 GBBM01001298 JAC34120.1 GL888609 EGI59050.1 AJFE02119515 AJFE02119516 AJFE02119517 AJFE02119518 AJFE02119519 AJFE02119520 AJFE02119521 AJFE02119522 AJFE02119523 AJFE02119524 AANG04003966 KQ976509 KYM82680.1 GFAC01005203 JAT93985.1 GDRN01076373 JAI62903.1 GANO01003514 JAB56357.1 APGK01043031 KB741011 KB631636 KI209682 ENN75554.1 ERL84883.1 ERL96074.1 CAEY01000714 GEBF01003892 JAN99740.1 AXCM01001816 ATLV01013353 KE524854 KFB37732.1 IACL01015259 IACL01015260 LAB00378.1 IACK01081177 IACK01081181 LAA79303.1 AADN05001052 AJ720674 CAG32333.1 KZ288266 PBC30267.1 GGFK01012372 MBW45693.1 JH171133 EHB10892.1 AGTP01010763 AGTP01010764 AGTP01010765 AGTP01010766 AGTP01010767 AGTP01010768 AGTP01010769 AGTP01010770 AGTP01010771 AGTP01010772 KQ977151 KYN05286.1 ADMH02000317 ETN67004.1 APCN01002367
Proteomes
UP000005204
UP000053240
UP000218220
UP000007151
UP000037510
UP000192223
+ More
UP000007266 UP000053105 UP000002358 UP000245037 UP000215335 UP000053097 UP000279307 UP000027135 UP000053825 UP000076502 UP000000311 UP000008237 UP000036403 UP000233020 UP000233180 UP000233200 UP000233080 UP000078492 UP000001811 UP000002277 UP000001595 UP000001519 UP000005640 UP000001073 UP000233140 UP000233220 UP000233120 UP000005205 UP000233060 UP000233100 UP000028761 UP000006718 UP000078541 UP000076408 UP000008225 UP000007755 UP000240080 UP000265300 UP000011712 UP000075920 UP000078540 UP000018467 UP000019118 UP000030742 UP000015104 UP000081671 UP000233160 UP000075883 UP000075900 UP000030765 UP000001645 UP000261540 UP000075885 UP000075880 UP000000539 UP000242457 UP000006813 UP000245320 UP000005215 UP000078542 UP000000673 UP000248482 UP000291020 UP000076407 UP000075903 UP000075882 UP000075902 UP000075840
UP000007266 UP000053105 UP000002358 UP000245037 UP000215335 UP000053097 UP000279307 UP000027135 UP000053825 UP000076502 UP000000311 UP000008237 UP000036403 UP000233020 UP000233180 UP000233200 UP000233080 UP000078492 UP000001811 UP000002277 UP000001595 UP000001519 UP000005640 UP000001073 UP000233140 UP000233220 UP000233120 UP000005205 UP000233060 UP000233100 UP000028761 UP000006718 UP000078541 UP000076408 UP000008225 UP000007755 UP000240080 UP000265300 UP000011712 UP000075920 UP000078540 UP000018467 UP000019118 UP000030742 UP000015104 UP000081671 UP000233160 UP000075883 UP000075900 UP000030765 UP000001645 UP000261540 UP000075885 UP000075880 UP000000539 UP000242457 UP000006813 UP000245320 UP000005215 UP000078542 UP000000673 UP000248482 UP000291020 UP000076407 UP000075903 UP000075882 UP000075902 UP000075840
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JTL8
A0A1E1WSF9
A0A194R1C3
A0A2A4JD18
S4PY51
A0A2H1VIX2
+ More
A0A212FN00 A0A0L7L2A4 A0A0L7LG43 A0A0L7L263 A0A1W4WUS4 D6WHX1 A0A0N0BKJ7 K7IP16 A0A2P8XS51 A0A232EUT9 A0A026WUS6 A0A293MT60 A0A2R5LD81 A0A0C9QSG8 A0A067RI08 A0A0L7QMR3 A0A154PF75 L7LQW8 E2A273 A0A1Z5L1F1 E2BNL7 A0A0J7K4Y7 G3MI98 A0A2K5ERL1 A0A224Z8T2 A0A2K6KKH5 A0A2K6NBV1 A0A2K5I8S4 A0A151IVP8 G1T7P0 E9IWP1 K7DEW9 A0A2J8Y4X1 G3RFB2 A0A024RD67 O75486 A0A2I3HJM7 A0A2K5ZLH1 A0A2K6V6B1 A0A2K6BFJ1 A0A158NN48 A0A2K5LG50 A0A2K5UI13 A0A2I3LH29 H9G0V2 A0A195FGS8 A0A131YW77 A0A182YJC1 Q5U608 U3D6N9 A0A023GML6 F4X369 A0A2R9AIQ8 A0A340XGK0 M3WBP7 A0A182WEJ1 A0A195BET6 A0A1E1X3Z4 A0A3B1KG57 A0A0P4W7L0 U5EGB3 N6T5T7 T1KYN1 A0A1S3FDC8 A0A2K6EMP3 A0A0P6J671 A0A182M2Q8 A0A182R6Z9 A0A084VID8 A0A2D4JV86 G3UR77 A0A3B3T3S8 A0A182PC97 A0A2D4I4Z9 A0A182J891 Q5ZIW0 A0A3Q2UIL6 A0A2A3EEV6 A0A2M4AY25 G5BNF1 A0A2R8MX91 A0A2U4BIY3 I3MPQ3 A0A151IKZ8 W5JVU5 A0A2Y9JIB1 A0A452J2P7 A0A182XKL5 A0A182VDM6 A0A182L4B0 A0A182TN70 A0A182HGX0
A0A212FN00 A0A0L7L2A4 A0A0L7LG43 A0A0L7L263 A0A1W4WUS4 D6WHX1 A0A0N0BKJ7 K7IP16 A0A2P8XS51 A0A232EUT9 A0A026WUS6 A0A293MT60 A0A2R5LD81 A0A0C9QSG8 A0A067RI08 A0A0L7QMR3 A0A154PF75 L7LQW8 E2A273 A0A1Z5L1F1 E2BNL7 A0A0J7K4Y7 G3MI98 A0A2K5ERL1 A0A224Z8T2 A0A2K6KKH5 A0A2K6NBV1 A0A2K5I8S4 A0A151IVP8 G1T7P0 E9IWP1 K7DEW9 A0A2J8Y4X1 G3RFB2 A0A024RD67 O75486 A0A2I3HJM7 A0A2K5ZLH1 A0A2K6V6B1 A0A2K6BFJ1 A0A158NN48 A0A2K5LG50 A0A2K5UI13 A0A2I3LH29 H9G0V2 A0A195FGS8 A0A131YW77 A0A182YJC1 Q5U608 U3D6N9 A0A023GML6 F4X369 A0A2R9AIQ8 A0A340XGK0 M3WBP7 A0A182WEJ1 A0A195BET6 A0A1E1X3Z4 A0A3B1KG57 A0A0P4W7L0 U5EGB3 N6T5T7 T1KYN1 A0A1S3FDC8 A0A2K6EMP3 A0A0P6J671 A0A182M2Q8 A0A182R6Z9 A0A084VID8 A0A2D4JV86 G3UR77 A0A3B3T3S8 A0A182PC97 A0A2D4I4Z9 A0A182J891 Q5ZIW0 A0A3Q2UIL6 A0A2A3EEV6 A0A2M4AY25 G5BNF1 A0A2R8MX91 A0A2U4BIY3 I3MPQ3 A0A151IKZ8 W5JVU5 A0A2Y9JIB1 A0A452J2P7 A0A182XKL5 A0A182VDM6 A0A182L4B0 A0A182TN70 A0A182HGX0
PDB
6MZL
E-value=0.00519473,
Score=93
Ontologies
GO
GO:0006366
GO:0046982
GO:0003712
GO:0003677
GO:0005669
GO:0006357
GO:0016740
GO:0033276
GO:0016578
GO:0004402
GO:0043966
GO:0030914
GO:0003713
GO:0005654
GO:0005634
GO:0006355
GO:0045893
GO:0003723
GO:0000178
GO:0006396
GO:0003743
GO:0008270
GO:0003676
GO:0005506
GO:0016705
GO:0020037
GO:0051259
GO:0008449
GO:0030203
GO:0006730
GO:0009258
GO:0016155
GO:0003824
Topology
Subcellular location
Nucleus
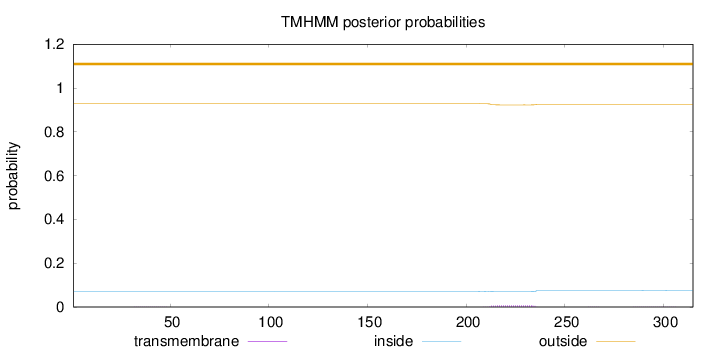
Length:
315
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.18369
Exp number, first 60 AAs:
0.00329
Total prob of N-in:
0.07175
outside
1 - 315
Population Genetic Test Statistics
Pi
287.256169
Theta
212.839416
Tajima's D
1.100805
CLR
0
CSRT
0.690565471726414
Interpretation
Uncertain
