Gene
KWMTBOMO09689 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012952
Annotation
N-acetylglucosamine-6-sulfatase_[Papilio_xuthus]
Full name
N-acetylglucosamine-6-sulfatase
Alternative Name
Glucosamine-6-sulfatase
Location in the cell
Extracellular Reliability : 1.639
Sequence
CDS
ATGTTACAATATTTATTTTTAATATTTTTCGTGAACAATGCTGTGGCTGAACTAAAACGACCAAACTTTGTTCTCATATTGACCGACGATCAAGATGTAGTTTTAGGTGGAATGGATCCGATGAAGAATGTTCAACGATTTATCGGGAAAGAAGGCATAACATTCACTAACTCCTACGTGACGTCACCGATATGCTGTCCAAGCAGAGCCAGTTTGCTGACAGGCATGTACGTTCACAACCACAAGACCGTGAACAATAGTCTTCACGGCGGATGCTATGGAGAGAACTGGAAAGATCACGAGAAACAGACGTTCGCTACTATACTTCAGGAAGCCGGATATGACACGTTCTATGCGGGGAAATATTTGAATCAGTATGGTACTAAAGAAGCCGGTGGCCCAGAGGTGGTACCCCCGGGCTGGACAGAATGGCGCGGCTTGGTCGGGAATTCAGTTTACTACAACTATACTTTGTCCAACAACGGCGTGCCGACGTTTTCGACTGATAGCTACTTAACCGATGTGATACGTGAACTGGGAGTCAGCTACATAGAGAATCAGACGGAATCGAAGCCCTTTCTGATGGTGCTCGCTCCCCCCGCCCCGCACCAGCCCTTCACCCCCGCCCCTCGACACAAAGACGCGTTCAGTAACGTGACAGTCGTAAAGAGCCCCAATTTCAATATCATCGCTGAGGATAAGCATTGGCTGCTCCGCATGCCTCCGTCGCCACTCCCTGACGTGATGCTTCCCGAACTCGACCAAGTGTACCGGTCCCGGTGGGAAACTCTGCTCTCGGTCGATGAGATGGTCGCCGACATTATAGGCGCTTTGGAAACTCAAGCGTTCCTGGATAACACATACGTAGTGTTCACTTCCGATAACGGTTACCATATCGGTCAAAACTCTCAAGTTTACGACAAACGTCAACCTTACGAAACTGACATCAAAGTTCCTTTACTAATAAAAGGGCCAGGACTGAAAAAGAACGTGGAAAACCGACAGCCGGTGTTAAATATTGATTTGGCGCCGACTATAATCAAACTGGCCGGCTTGGACATACCGAACAACGTGGACGGAAGGCCCATCGATGTTCACAACGAGGACATAGGCGAGAGGAACATGCTAATTGAATACTACGGCGAAGGCAAGGACGGATCTGTGGATAAAGAATGTCCTTGGAAGTATGATAGCGGTAATCTCGCTCAATGCTATCCTCAGTACAGTTGCAAGTGTCAAGACGCCAAAAACAATACATACGCCTGTATCAGACACATCGCGGAGCGAATCGACAAGAAATATTGCCTCTTCGCTGACAATGAGAACTTCAGTGAAATGTACAACATAGCAGACGATCCTTACGAATTGGAGAACATTGTGAACCACGTGTTTCCGGCGGTGAAGTACTGGTACAGAGAGATTCTAACGCGGATGCTGACCTGCTCCGGTGCTCATAATTGTGATCACCCATTGGAACCACTAATTAATATATTTTAG
Protein
MLQYLFLIFFVNNAVAELKRPNFVLILTDDQDVVLGGMDPMKNVQRFIGKEGITFTNSYVTSPICCPSRASLLTGMYVHNHKTVNNSLHGGCYGENWKDHEKQTFATILQEAGYDTFYAGKYLNQYGTKEAGGPEVVPPGWTEWRGLVGNSVYYNYTLSNNGVPTFSTDSYLTDVIRELGVSYIENQTESKPFLMVLAPPAPHQPFTPAPRHKDAFSNVTVVKSPNFNIIAEDKHWLLRMPPSPLPDVMLPELDQVYRSRWETLLSVDEMVADIIGALETQAFLDNTYVVFTSDNGYHIGQNSQVYDKRQPYETDIKVPLLIKGPGLKKNVENRQPVLNIDLAPTIIKLAGLDIPNNVDGRPIDVHNEDIGERNMLIEYYGEGKDGSVDKECPWKYDSGNLAQCYPQYSCKCQDAKNNTYACIRHIAERIDKKYCLFADNENFSEMYNIADDPYELENIVNHVFPAVKYWYREILTRMLTCSGAHNCDHPLEPLINIF
Summary
Catalytic Activity
Hydrolysis of the 6-sulfate groups of the N-acetyl-D-glucosamine 6-sulfate units of heparan sulfate and keratan sulfate.
Cofactor
Ca(2+)
Similarity
Belongs to the sulfatase family.
Feature
chain N-acetylglucosamine-6-sulfatase
Uniprot
H9JTU0
A0A3S2L6R0
A0A194Q0P5
A0A194R227
S4PAQ9
A0A212FN11
+ More
A0A2A4JY89 A0A2H1WZ35 A0A067R287 A0A2M4ATL5 A0A2M4ATQ1 A0A2M4BL80 A0A2M4BME1 Q17CP8 A0A182NN85 A0A2M3ZID9 A0A2M4BLJ4 A0A1W7R8K1 A0A023EVZ0 A0A182QZW2 A0A2M4DKR4 A0A2M4DKP3 A0A182R2T8 A0A1Q3FJH7 A0A034VMM1 A0A0K8VY72 A0A2J7R0V6 A0A182JMF2 A0A182K5F3 A0A2M3Z9R5 Q7Q2M5 A0A151JUB5 A0A0L0CP35 A0A232F1L4 W8B4N7 A0A1B6CPK4 A0A088A3D6 A0A1A9WMW8 R7TJ59 E9IVX7 E0VPN4 A0A336K4J7 T1PJN0 A0A1I8N5Y6 A0A2A3EDB1 A0A1A9XVG4 A0A3Q2Z177 A0A1B0AXE5 A0A154PS62 A0A1A9V2D5 A0A0A1WVI1 B3NRL5 A0A1B6KZS0 A0A0J9RCY8 A0A1A9ZYD8 Q5BIL9 B4GCV2 B4HQI3 E2ANB3 B4MIX0 A0A0C9QER4 B4JJV2 B5E066 Q95R73 A0A3B4VQF6 Q86NX7 A0A0P4WDR7 A0A1W4V529 B4P4I0 A0A3Q4M983 A0A3P8PYQ7 B4L7H5 A0A3P9C342 A0A2L2YGX1 H2YLW8 A0A3B4Y3C4 A0A182T8A8 A0A401PMW9 A0A3Q2WM37 A0A310SFM3 A0A1L8GYG5 A0A401RWF2 A0A3B4FRV1 A0A3Q3LPY1 A0A087YLI7 A0A3P8S7Z9 A0A3B3VQE4 V9K9M2 A0A401P548 A0A3Q1C3T3 A0A1Q3FJG5 A0A3N0Z2X4 A9JRM2 A0A3B0IZM1 B4MGI5 S4RF84 A0A3B3YC40 A0A3Q3GD06 A0A3P9PLX2 A0A401RY68
A0A2A4JY89 A0A2H1WZ35 A0A067R287 A0A2M4ATL5 A0A2M4ATQ1 A0A2M4BL80 A0A2M4BME1 Q17CP8 A0A182NN85 A0A2M3ZID9 A0A2M4BLJ4 A0A1W7R8K1 A0A023EVZ0 A0A182QZW2 A0A2M4DKR4 A0A2M4DKP3 A0A182R2T8 A0A1Q3FJH7 A0A034VMM1 A0A0K8VY72 A0A2J7R0V6 A0A182JMF2 A0A182K5F3 A0A2M3Z9R5 Q7Q2M5 A0A151JUB5 A0A0L0CP35 A0A232F1L4 W8B4N7 A0A1B6CPK4 A0A088A3D6 A0A1A9WMW8 R7TJ59 E9IVX7 E0VPN4 A0A336K4J7 T1PJN0 A0A1I8N5Y6 A0A2A3EDB1 A0A1A9XVG4 A0A3Q2Z177 A0A1B0AXE5 A0A154PS62 A0A1A9V2D5 A0A0A1WVI1 B3NRL5 A0A1B6KZS0 A0A0J9RCY8 A0A1A9ZYD8 Q5BIL9 B4GCV2 B4HQI3 E2ANB3 B4MIX0 A0A0C9QER4 B4JJV2 B5E066 Q95R73 A0A3B4VQF6 Q86NX7 A0A0P4WDR7 A0A1W4V529 B4P4I0 A0A3Q4M983 A0A3P8PYQ7 B4L7H5 A0A3P9C342 A0A2L2YGX1 H2YLW8 A0A3B4Y3C4 A0A182T8A8 A0A401PMW9 A0A3Q2WM37 A0A310SFM3 A0A1L8GYG5 A0A401RWF2 A0A3B4FRV1 A0A3Q3LPY1 A0A087YLI7 A0A3P8S7Z9 A0A3B3VQE4 V9K9M2 A0A401P548 A0A3Q1C3T3 A0A1Q3FJG5 A0A3N0Z2X4 A9JRM2 A0A3B0IZM1 B4MGI5 S4RF84 A0A3B3YC40 A0A3Q3GD06 A0A3P9PLX2 A0A401RY68
EC Number
3.1.6.14
Pubmed
19121390
26354079
23622113
22118469
24845553
17510324
+ More
24945155 25348373 12364791 14747013 17210077 26108605 28648823 24495485 23254933 21282665 20566863 25315136 25830018 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 15632085 17550304 25186727 26561354 30297745 27762356 24402279 18057021
24945155 25348373 12364791 14747013 17210077 26108605 28648823 24495485 23254933 21282665 20566863 25315136 25830018 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 15632085 17550304 25186727 26561354 30297745 27762356 24402279 18057021
EMBL
BABH01003609
BABH01003610
RSAL01000115
RVE46899.1
KQ459582
KPI98878.1
+ More
KQ460878 KPJ11569.1 GAIX01004896 JAA87664.1 AGBW02007649 OWR55125.1 NWSH01000362 PCG76985.1 ODYU01012168 SOQ58339.1 KK852979 KDR13030.1 GGFK01010796 MBW44117.1 GGFK01010826 MBW44147.1 GGFJ01004685 MBW53826.1 GGFJ01004797 MBW53938.1 CH477305 EAT44148.1 GGFM01007528 MBW28279.1 GGFJ01004796 MBW53937.1 GEHC01000178 JAV47467.1 GAPW01001109 JAC12489.1 AXCN02001899 GGFL01013946 MBW78124.1 GGFL01013945 MBW78123.1 GFDL01007402 JAV27643.1 GAKP01015268 GAKP01015266 JAC43684.1 GDHF01008530 JAI43784.1 NEVH01008217 PNF34456.1 GGFM01004513 MBW25264.1 AAAB01008968 EAA13233.3 KQ981799 KYN35674.1 JRES01000104 KNC34095.1 NNAY01001304 OXU24419.1 GAMC01014527 JAB92028.1 GEDC01021892 JAS15406.1 AMQN01013733 KB310433 ELT91586.1 GL766449 EFZ15247.1 DS235366 EEB15340.1 UFQS01000022 UFQT01000022 SSW97623.1 SSX18009.1 KA648153 AFP62782.1 KZ288293 PBC29011.1 JXJN01005238 KQ435026 KZC13950.1 GBXI01011651 JAD02641.1 CH954179 EDV56167.1 GEBQ01023024 JAT16953.1 CM002911 KMY93479.1 AE013599 BT021205 AAG22274.2 AAX33353.1 CH479181 EDW32515.1 CH480816 EDW47718.1 GL441177 EFN65061.1 CH963719 EDW72059.2 GBYB01013013 JAG82780.1 CH916370 EDV99854.1 CM000071 EDY69300.2 AY061585 AAL29133.1 AAM68572.1 BT003587 AAO39590.1 GDRN01060069 JAI65436.1 CM000158 EDW90619.1 CH933813 EDW10969.1 IAAA01024018 LAA07394.1 BFAA01000980 GCB74459.1 KQ760495 OAD60118.1 CM004470 OCT88892.1 BEZZ01000011 GCC22476.1 AYCK01014435 AYCK01014436 JW861953 AFO94470.1 BFAA01005864 GCB68264.1 GFDL01007381 JAV27664.1 RJVU01015140 ROL52603.1 BC155714 AAI55715.1 OUUW01000001 SPP73874.1 CH940677 EDW58302.1 KRF78210.1 KRF78211.1 BEZZ01000022 GCC23049.1
KQ460878 KPJ11569.1 GAIX01004896 JAA87664.1 AGBW02007649 OWR55125.1 NWSH01000362 PCG76985.1 ODYU01012168 SOQ58339.1 KK852979 KDR13030.1 GGFK01010796 MBW44117.1 GGFK01010826 MBW44147.1 GGFJ01004685 MBW53826.1 GGFJ01004797 MBW53938.1 CH477305 EAT44148.1 GGFM01007528 MBW28279.1 GGFJ01004796 MBW53937.1 GEHC01000178 JAV47467.1 GAPW01001109 JAC12489.1 AXCN02001899 GGFL01013946 MBW78124.1 GGFL01013945 MBW78123.1 GFDL01007402 JAV27643.1 GAKP01015268 GAKP01015266 JAC43684.1 GDHF01008530 JAI43784.1 NEVH01008217 PNF34456.1 GGFM01004513 MBW25264.1 AAAB01008968 EAA13233.3 KQ981799 KYN35674.1 JRES01000104 KNC34095.1 NNAY01001304 OXU24419.1 GAMC01014527 JAB92028.1 GEDC01021892 JAS15406.1 AMQN01013733 KB310433 ELT91586.1 GL766449 EFZ15247.1 DS235366 EEB15340.1 UFQS01000022 UFQT01000022 SSW97623.1 SSX18009.1 KA648153 AFP62782.1 KZ288293 PBC29011.1 JXJN01005238 KQ435026 KZC13950.1 GBXI01011651 JAD02641.1 CH954179 EDV56167.1 GEBQ01023024 JAT16953.1 CM002911 KMY93479.1 AE013599 BT021205 AAG22274.2 AAX33353.1 CH479181 EDW32515.1 CH480816 EDW47718.1 GL441177 EFN65061.1 CH963719 EDW72059.2 GBYB01013013 JAG82780.1 CH916370 EDV99854.1 CM000071 EDY69300.2 AY061585 AAL29133.1 AAM68572.1 BT003587 AAO39590.1 GDRN01060069 JAI65436.1 CM000158 EDW90619.1 CH933813 EDW10969.1 IAAA01024018 LAA07394.1 BFAA01000980 GCB74459.1 KQ760495 OAD60118.1 CM004470 OCT88892.1 BEZZ01000011 GCC22476.1 AYCK01014435 AYCK01014436 JW861953 AFO94470.1 BFAA01005864 GCB68264.1 GFDL01007381 JAV27664.1 RJVU01015140 ROL52603.1 BC155714 AAI55715.1 OUUW01000001 SPP73874.1 CH940677 EDW58302.1 KRF78210.1 KRF78211.1 BEZZ01000022 GCC23049.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000053240
UP000007151
UP000218220
+ More
UP000027135 UP000008820 UP000075884 UP000075886 UP000075900 UP000235965 UP000075880 UP000075881 UP000007062 UP000078541 UP000037069 UP000215335 UP000005203 UP000091820 UP000014760 UP000009046 UP000095301 UP000242457 UP000092443 UP000264820 UP000092460 UP000076502 UP000078200 UP000008711 UP000092445 UP000000803 UP000008744 UP000001292 UP000000311 UP000007798 UP000001070 UP000001819 UP000261420 UP000192221 UP000002282 UP000261580 UP000265100 UP000009192 UP000265160 UP000007875 UP000261360 UP000075901 UP000288216 UP000264840 UP000186698 UP000287033 UP000261460 UP000261640 UP000028760 UP000265080 UP000261500 UP000257160 UP000268350 UP000008792 UP000245300 UP000261480 UP000261660 UP000242638
UP000027135 UP000008820 UP000075884 UP000075886 UP000075900 UP000235965 UP000075880 UP000075881 UP000007062 UP000078541 UP000037069 UP000215335 UP000005203 UP000091820 UP000014760 UP000009046 UP000095301 UP000242457 UP000092443 UP000264820 UP000092460 UP000076502 UP000078200 UP000008711 UP000092445 UP000000803 UP000008744 UP000001292 UP000000311 UP000007798 UP000001070 UP000001819 UP000261420 UP000192221 UP000002282 UP000261580 UP000265100 UP000009192 UP000265160 UP000007875 UP000261360 UP000075901 UP000288216 UP000264840 UP000186698 UP000287033 UP000261460 UP000261640 UP000028760 UP000265080 UP000261500 UP000257160 UP000268350 UP000008792 UP000245300 UP000261480 UP000261660 UP000242638
Pfam
PF00884 Sulfatase
Interpro
SUPFAM
SSF53649
SSF53649
Gene 3D
ProteinModelPortal
H9JTU0
A0A3S2L6R0
A0A194Q0P5
A0A194R227
S4PAQ9
A0A212FN11
+ More
A0A2A4JY89 A0A2H1WZ35 A0A067R287 A0A2M4ATL5 A0A2M4ATQ1 A0A2M4BL80 A0A2M4BME1 Q17CP8 A0A182NN85 A0A2M3ZID9 A0A2M4BLJ4 A0A1W7R8K1 A0A023EVZ0 A0A182QZW2 A0A2M4DKR4 A0A2M4DKP3 A0A182R2T8 A0A1Q3FJH7 A0A034VMM1 A0A0K8VY72 A0A2J7R0V6 A0A182JMF2 A0A182K5F3 A0A2M3Z9R5 Q7Q2M5 A0A151JUB5 A0A0L0CP35 A0A232F1L4 W8B4N7 A0A1B6CPK4 A0A088A3D6 A0A1A9WMW8 R7TJ59 E9IVX7 E0VPN4 A0A336K4J7 T1PJN0 A0A1I8N5Y6 A0A2A3EDB1 A0A1A9XVG4 A0A3Q2Z177 A0A1B0AXE5 A0A154PS62 A0A1A9V2D5 A0A0A1WVI1 B3NRL5 A0A1B6KZS0 A0A0J9RCY8 A0A1A9ZYD8 Q5BIL9 B4GCV2 B4HQI3 E2ANB3 B4MIX0 A0A0C9QER4 B4JJV2 B5E066 Q95R73 A0A3B4VQF6 Q86NX7 A0A0P4WDR7 A0A1W4V529 B4P4I0 A0A3Q4M983 A0A3P8PYQ7 B4L7H5 A0A3P9C342 A0A2L2YGX1 H2YLW8 A0A3B4Y3C4 A0A182T8A8 A0A401PMW9 A0A3Q2WM37 A0A310SFM3 A0A1L8GYG5 A0A401RWF2 A0A3B4FRV1 A0A3Q3LPY1 A0A087YLI7 A0A3P8S7Z9 A0A3B3VQE4 V9K9M2 A0A401P548 A0A3Q1C3T3 A0A1Q3FJG5 A0A3N0Z2X4 A9JRM2 A0A3B0IZM1 B4MGI5 S4RF84 A0A3B3YC40 A0A3Q3GD06 A0A3P9PLX2 A0A401RY68
A0A2A4JY89 A0A2H1WZ35 A0A067R287 A0A2M4ATL5 A0A2M4ATQ1 A0A2M4BL80 A0A2M4BME1 Q17CP8 A0A182NN85 A0A2M3ZID9 A0A2M4BLJ4 A0A1W7R8K1 A0A023EVZ0 A0A182QZW2 A0A2M4DKR4 A0A2M4DKP3 A0A182R2T8 A0A1Q3FJH7 A0A034VMM1 A0A0K8VY72 A0A2J7R0V6 A0A182JMF2 A0A182K5F3 A0A2M3Z9R5 Q7Q2M5 A0A151JUB5 A0A0L0CP35 A0A232F1L4 W8B4N7 A0A1B6CPK4 A0A088A3D6 A0A1A9WMW8 R7TJ59 E9IVX7 E0VPN4 A0A336K4J7 T1PJN0 A0A1I8N5Y6 A0A2A3EDB1 A0A1A9XVG4 A0A3Q2Z177 A0A1B0AXE5 A0A154PS62 A0A1A9V2D5 A0A0A1WVI1 B3NRL5 A0A1B6KZS0 A0A0J9RCY8 A0A1A9ZYD8 Q5BIL9 B4GCV2 B4HQI3 E2ANB3 B4MIX0 A0A0C9QER4 B4JJV2 B5E066 Q95R73 A0A3B4VQF6 Q86NX7 A0A0P4WDR7 A0A1W4V529 B4P4I0 A0A3Q4M983 A0A3P8PYQ7 B4L7H5 A0A3P9C342 A0A2L2YGX1 H2YLW8 A0A3B4Y3C4 A0A182T8A8 A0A401PMW9 A0A3Q2WM37 A0A310SFM3 A0A1L8GYG5 A0A401RWF2 A0A3B4FRV1 A0A3Q3LPY1 A0A087YLI7 A0A3P8S7Z9 A0A3B3VQE4 V9K9M2 A0A401P548 A0A3Q1C3T3 A0A1Q3FJG5 A0A3N0Z2X4 A9JRM2 A0A3B0IZM1 B4MGI5 S4RF84 A0A3B3YC40 A0A3Q3GD06 A0A3P9PLX2 A0A401RY68
PDB
3ED4
E-value=2.42707e-17,
Score=219
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Lysosome
SignalP
Position: 1 - 16,
Likelihood: 0.864498
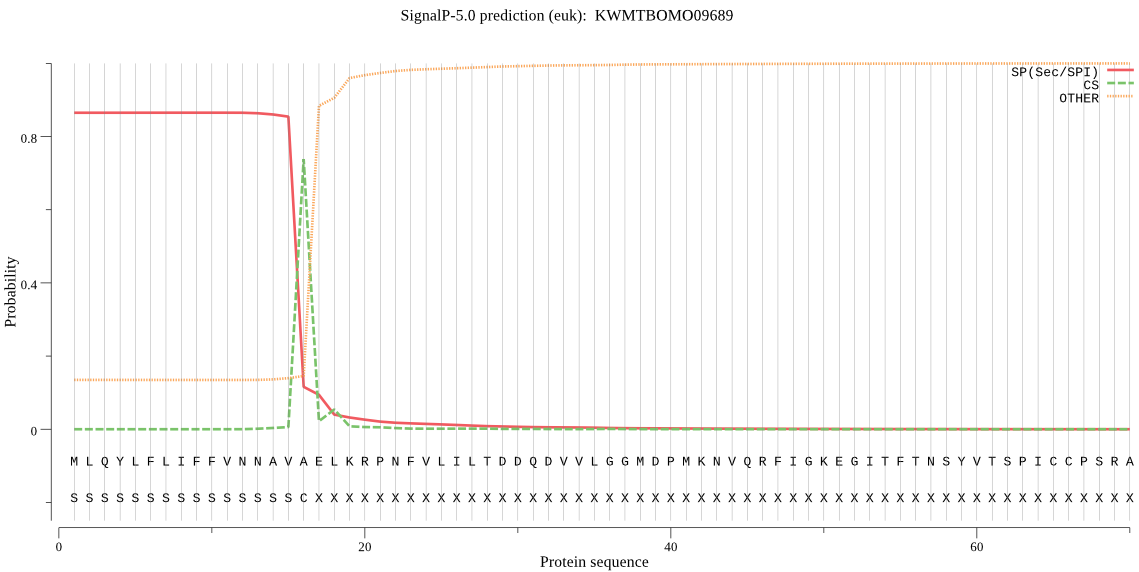
Length:
498
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00227
Exp number, first 60 AAs:
0.00203
Total prob of N-in:
0.00023
outside
1 - 498
Population Genetic Test Statistics
Pi
231.224388
Theta
159.311841
Tajima's D
1.521154
CLR
0.195791
CSRT
0.792410379481026
Interpretation
Uncertain
