Pre Gene Modal
BGIBMGA012953
Annotation
PREDICTED:_vacuole_membrane_protein_1_isoform_X1_[Bombyx_mori]
Full name
Vacuole membrane protein 1
Alternative Name
NF-E2-inducible protein 2
Transmembrane protein 49
Transmembrane protein 49
Location in the cell
PlasmaMembrane Reliability : 3.011
Sequence
CDS
ATGGCAGGCACAAGACGTGCACGTAATCAGGTGAATTCGCTGCCTATGCCTCGTCAGAATTCAGTGTCTGACAATGGGCGAGCACGCCAGGGCCTGAGCTCAGAAGCTTTAGCCGCTATGACTAAGGAACAGCTACGTGCAGAGTGTCGGAAAAGAGGTCAACGTGCAACGGGAAACAAAAACGAGCTGCTCGCCCGTCTCGGGTATTACAAGCTGGCGGTCACTCCTCAGCCTCAGGACAGCGAACAGCGCATGCGCAACGGCTTGCACGCTCCACACCCGAAGGAGATAAGGACCAAAACAAATTCCTCAATGGAGGCCGACAGTCTTGTGTTGTGGAGGCGGCCCCTCATCACATTGGAGTATTTCTTCAGAGAACTTTTTATATTAATCAACACAGGATTGCAGAGATTGATTGCATACAGAGTACTGGCGTTAACAATAGCCGTGGCAATAATTGGTACTGCGATTTCGTATTATATCAGCGGCCCTCATCAGCAGTACGTGCATTTAGTTGTAGCCACTCTAGCCTGGTGGGGCTGGTGGGTAGTTCTAGGCGTTTGTAGTTCTGTGGGCCTCGGTACGGGCTTGCACACATTCCTATTGTATTTGGGGCCGCACATAGCGAGGGTTACACTCGCAGCGTATGAATGTGGAGGACTAAACTTCCCGGAGCCACCTTATCCTAATGATATAATTTGTCCGGCGGAGGTCGATCCTAACTACGCGGTATCGATTTGGAATATAATGGCGAAAGTTCGCGTCGAATCCATGATGTGGGGCATAGGCACCGCTCTAGGCGAACTGCCGCCGTACTTCATGGCGAGGGCCGCTAGACTCTCAGGGGCCGGGGTCGCCGAACTCAACACCGACGATGATTCGAGGACCGGAAGGGCTAAAATAATGGTGCAGAAATTAGTGCAGCGCGTAGGCTTTGCTGGTATATTGGCTTGCGCTTCGGTGCCTAACCCTCTCTTCGATTTGGCCGGCCTGACCTGCGGACACTTCTTGGTGCCGTTTTGGACGTTCTTCGGAGCGACGGTCATCGGGAAAGCGGTCATAAAGATGCACCTGCAGAAAATGTTCGTCATTATCGCGTTCAACGAGACTTTGGTTGGGCAAGCGTTGTCTTGGGTCGAAAAAATACCGTACATCGGTCCGAAATTAGAGGCTCCGCTTTTGGAATTCTTGAGGAACCAGAAAGCGCGTCTTCACAAGAACGATTCCACGCAGATACCAGAAAATCAGGGTTCGGTATTATCGAACATTCTAGAAAAGTTCGTTCTGGCGATGGTCGTTTACTTTATTGTGTCGATTGTGAACGCTTTGGCCCAAAACTACAGTAAGAGGGTCGGTAAGAAGAAGAGCAAGAAAAGAGAATAG
Protein
MAGTRRARNQVNSLPMPRQNSVSDNGRARQGLSSEALAAMTKEQLRAECRKRGQRATGNKNELLARLGYYKLAVTPQPQDSEQRMRNGLHAPHPKEIRTKTNSSMEADSLVLWRRPLITLEYFFRELFILINTGLQRLIAYRVLALTIAVAIIGTAISYYISGPHQQYVHLVVATLAWWGWWVVLGVCSSVGLGTGLHTFLLYLGPHIARVTLAAYECGGLNFPEPPYPNDIICPAEVDPNYAVSIWNIMAKVRVESMMWGIGTALGELPPYFMARAARLSGAGVAELNTDDDSRTGRAKIMVQKLVQRVGFAGILACASVPNPLFDLAGLTCGHFLVPFWTFFGATVIGKAVIKMHLQKMFVIIAFNETLVGQALSWVEKIPYIGPKLEAPLLEFLRNQKARLHKNDSTQIPENQGSVLSNILEKFVLAMVVYFIVSIVNALAQNYSKRVGKKKSKKRE
Summary
Description
Stress-induced protein that, when overexpressed, promotes formation of intracellular vacuoles followed by cell death (By similarity). May be involved in the cytoplasmic vacuolization of acinar cells during the early stage of acute pancreatitis (PubMed:17940279). Involved in cell-cell adhesion. Plays an essential role in formation of cell junctions. Plays a role in the initial stages of the autophagic process through its interaction with BECN1. Required for autophagosome formation (By similarity).
Stress-induced protein that, when overexpressed, promotes formation of intracellular vacuoles followed by cell death (PubMed:11785947). May be involved in the cytoplasmic vacuolization of acinar cells during the early stage of acute pancreatitis (PubMed:12649568, PubMed:15367889). Involved in cell-cell adhesion (By similarity). Plays an essential role in formation of cell junctions (By similarity). Plays a role in the initial stages of the autophagic process through its interaction with BECN1 (PubMed:17940279). Required for autophagosome formation (By similarity).
Stress-induced protein that, when overexpressed, promotes formation of intracellular vacuoles followed by cell death. May be involved in the cytoplasmic vacuolization of acinar cells during the early stage of acute pancreatitis. Involved in cell-cell adhesion. Plays an essential role in formation of cell junctions. Plays a role in the initial stages of the autophagic process through its interaction with BECN1. Required for autophagosome formation (By similarity).
Stress-induced protein that, when overexpressed, promotes formation of intracellular vacuoles followed by cell death (PubMed:11785947). May be involved in the cytoplasmic vacuolization of acinar cells during the early stage of acute pancreatitis (PubMed:12649568, PubMed:15367889). Involved in cell-cell adhesion (By similarity). Plays an essential role in formation of cell junctions (By similarity). Plays a role in the initial stages of the autophagic process through its interaction with BECN1 (PubMed:17940279). Required for autophagosome formation (By similarity).
Stress-induced protein that, when overexpressed, promotes formation of intracellular vacuoles followed by cell death. May be involved in the cytoplasmic vacuolization of acinar cells during the early stage of acute pancreatitis. Involved in cell-cell adhesion. Plays an essential role in formation of cell junctions. Plays a role in the initial stages of the autophagic process through its interaction with BECN1. Required for autophagosome formation (By similarity).
Subunit
Interacts with BECN1 (By similarity). Interacts with TJP1 (By similarity). Interacts with TP53INP2 (By similarity). Interacts with TMEM41B (By similarity).
Interacts with TJP1 (By similarity). Interacts with BECN1 (PubMed:17940279). Interacts with TP53INP2 (By similarity). Interacts with TMEM41B (By similarity).
Interacts with TJP1 (By similarity). Interacts with BECN1 (PubMed:17940279). Interacts with TP53INP2 (By similarity). Interacts with TMEM41B (By similarity).
Similarity
Belongs to the VMP1 family.
Keywords
Acetylation
Autophagy
Cell adhesion
Cell membrane
Complete proteome
Endoplasmic reticulum
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Vacuole
Feature
chain Vacuole membrane protein 1
Uniprot
A0A2A4JZY8
A0A212FBQ9
A0A194Q645
A0A194R1K1
A0A2H1VU66
A0A437B8N5
+ More
A0A2A4JY81 A0A1W4WRU7 A0A1Y1M5D5 D2A456 A0A067R8P9 N6SXI5 A0A1W4X2T3 A0A2J7PRE6 U4UPM4 A0A023EV67 A0A2P8Z4E6 Q0IEV8 A0A0P6IY74 T1DI36 A0A2J7PRF6 A0A1Y1M8U0 E0VMP9 A0A1Z5LHI1 J9K896 A0A131XC51 A0A2S2QVQ3 A0A1L8DUW3 A0A2R5LJT0 A0A2S2NIL2 L7MJ41 A0A1J1HWW3 A0A1J1HYV6 A0A0C9QV73 A0A1B6H9L4 J3JX80 A0A293M0N7 A0A3B3YLG3 V9KEE5 A0A3P9NVI1 A0A224ZB22 M3ZD62 A0A3B3ULI2 A0A131Z2A8 Q99KU0 A0A1I8P765 A0A2Y9T4G7 A0A2U4C5Y9 I3MR59 B7PTG6 F6PQC8 Q68EQ9 A0A1E1XSX9 A0A1B6FCN4 A0A2Y9T6B8 A0A2Y9DPJ9 A0A087Y456 A0A0P5ZT58 W5M8B4 A0A1L8H860 A0A087USE1 A0A0P6DET1 A0A2P6KEG0 A0A341CFG8 A0A2Y9LFD4 A0A0P4YUV6 A0A0P5A927 A0A401SRA5 A0A0P5XN04 A0A0P5M7J0 A0A340WK98 Q91ZQ0 A0A2U3Y9I3 A0A3Q2FW45 A0A2U3VRG2 A0A0P5HMK8 A0A3Q7MK72 A0A091DM61 A0A0P5SH55 A0A0P5J8B9 A0A0M4EMP9 A0A2Y9IBG3 A0A0P5KUU4 A0A401NUR4 A0A1U7UT36 S7P5P1 A0A2K5QG81 A0A2K6STR0 A0A2I3HZ10 G3TCW1 A0A2J8W7G8 Q5R9K4 F7EZ12 K7FP29 A0A1I8MIE4 A0A1S3AEJ0 G3QIS0 A0A2R9APT4 G2HG37
A0A2A4JY81 A0A1W4WRU7 A0A1Y1M5D5 D2A456 A0A067R8P9 N6SXI5 A0A1W4X2T3 A0A2J7PRE6 U4UPM4 A0A023EV67 A0A2P8Z4E6 Q0IEV8 A0A0P6IY74 T1DI36 A0A2J7PRF6 A0A1Y1M8U0 E0VMP9 A0A1Z5LHI1 J9K896 A0A131XC51 A0A2S2QVQ3 A0A1L8DUW3 A0A2R5LJT0 A0A2S2NIL2 L7MJ41 A0A1J1HWW3 A0A1J1HYV6 A0A0C9QV73 A0A1B6H9L4 J3JX80 A0A293M0N7 A0A3B3YLG3 V9KEE5 A0A3P9NVI1 A0A224ZB22 M3ZD62 A0A3B3ULI2 A0A131Z2A8 Q99KU0 A0A1I8P765 A0A2Y9T4G7 A0A2U4C5Y9 I3MR59 B7PTG6 F6PQC8 Q68EQ9 A0A1E1XSX9 A0A1B6FCN4 A0A2Y9T6B8 A0A2Y9DPJ9 A0A087Y456 A0A0P5ZT58 W5M8B4 A0A1L8H860 A0A087USE1 A0A0P6DET1 A0A2P6KEG0 A0A341CFG8 A0A2Y9LFD4 A0A0P4YUV6 A0A0P5A927 A0A401SRA5 A0A0P5XN04 A0A0P5M7J0 A0A340WK98 Q91ZQ0 A0A2U3Y9I3 A0A3Q2FW45 A0A2U3VRG2 A0A0P5HMK8 A0A3Q7MK72 A0A091DM61 A0A0P5SH55 A0A0P5J8B9 A0A0M4EMP9 A0A2Y9IBG3 A0A0P5KUU4 A0A401NUR4 A0A1U7UT36 S7P5P1 A0A2K5QG81 A0A2K6STR0 A0A2I3HZ10 G3TCW1 A0A2J8W7G8 Q5R9K4 F7EZ12 K7FP29 A0A1I8MIE4 A0A1S3AEJ0 G3QIS0 A0A2R9APT4 G2HG37
Pubmed
22118469
26354079
28004739
18362917
19820115
24845553
+ More
23537049 24945155 29403074 17510324 26999592 24330624 20566863 28528879 28049606 25576852 22516182 24402279 28797301 23542700 26830274 16141072 19468303 15489334 17940279 21183079 20431018 29209593 27762356 30297745 11785947 12649568 15367889 25243066 17381049 25315136 22398555 22722832 21484476
23537049 24945155 29403074 17510324 26999592 24330624 20566863 28528879 28049606 25576852 22516182 24402279 28797301 23542700 26830274 16141072 19468303 15489334 17940279 21183079 20431018 29209593 27762356 30297745 11785947 12649568 15367889 25243066 17381049 25315136 22398555 22722832 21484476
EMBL
NWSH01000362
PCG76980.1
AGBW02009293
OWR51170.1
KQ459582
KPI98880.1
+ More
KQ460878 KPJ11567.1 ODYU01004448 SOQ44328.1 RSAL01000115 RVE46897.1 PCG76977.1 GEZM01040026 JAV80992.1 KQ971348 EFA04843.1 KK852622 KDR19990.1 APGK01052855 KB741216 ENN72479.1 NEVH01022362 PNF18902.1 KB632404 ERL95062.1 GAPW01001224 JAC12374.1 PYGN01000201 PSN51347.1 CH477464 EAT40513.1 GDUN01001104 JAN94815.1 GALA01001171 JAA93681.1 PNF18899.1 GEZM01040028 JAV80990.1 DS235321 EEB14655.1 GFJQ02000088 JAW06882.1 ABLF02038839 GEFH01004042 JAP64539.1 GGMS01011999 MBY81202.1 GFDF01003875 JAV10209.1 GGLE01005552 MBY09678.1 GGMR01004376 MBY16995.1 GACK01001044 JAA63990.1 CVRI01000030 CRK92509.1 CRK92508.1 GBYB01004572 GBYB01012001 JAG74339.1 JAG81768.1 GECU01036271 JAS71435.1 BT127848 AEE62810.1 GFWV01009147 MAA33876.1 JW863603 AFO96120.1 GFPF01013267 MAA24413.1 GEDV01003625 JAP84932.1 AB047550 AB076609 AK043091 AK077443 AK159468 AL592222 AL604063 BC004013 AGTP01022571 AGTP01022572 AGTP01022573 AGTP01022574 AGTP01022575 AGTP01022576 AGTP01022577 AGTP01022578 AGTP01022579 AGTP01022580 ABJB010145156 ABJB010187107 ABJB010333299 ABJB010368120 ABJB010697092 DS785766 EEC09888.1 AAMC01027125 AAMC01027126 AAMC01027127 AAMC01027128 AAMC01027129 AAMC01027130 AAMC01027131 AAMC01027132 BC080142 GFAA01001026 JAU02409.1 GECZ01021818 JAS47951.1 AYCK01007209 AYCK01007210 AYCK01007211 AYCK01007212 AYCK01007213 GDIP01039654 JAM64061.1 AHAT01005716 AHAT01005717 CM004469 OCT92226.1 KK121348 KFM80280.1 GDIQ01078297 JAN16440.1 MWRG01014306 PRD24687.1 GDIP01223326 JAJ00076.1 GDIP01205824 GDIP01143042 LRGB01003071 JAJ17578.1 KZS04754.1 BEZZ01000472 GCC32917.1 GDIP01069753 JAM33962.1 GDIQ01159617 JAK92108.1 AF411216 BC061721 GDIQ01248190 JAK03535.1 KN122207 KFO32167.1 GDIP01139788 JAL63926.1 GDIQ01203662 JAK48063.1 CP012528 ALC49071.1 GDIQ01184271 JAK67454.1 BFAA01001385 GCB64631.1 KE161298 EPQ02932.1 ADFV01107744 ADFV01107745 ADFV01107746 ADFV01107747 ADFV01107748 ADFV01107749 ADFV01107750 ADFV01107751 ADFV01107752 ADFV01107753 NDHI03003397 PNJ65719.1 CR859383 GAMS01007511 GAMR01000421 GAMQ01003879 JAB15625.1 JAB33511.1 JAB37972.1 AGCU01199602 AGCU01199603 AGCU01199604 AGCU01199605 CABD030036879 CABD030036880 CABD030036881 CABD030036882 CABD030036883 CABD030036884 CABD030036885 CABD030036886 AJFE02058748 AJFE02058749 AJFE02058750 AJFE02058751 AJFE02058752 AJFE02058753 AJFE02058754 AJFE02058755 AJFE02058756 AJFE02058757 AK305701 GABC01004282 GABF01001689 GABD01004330 GABE01007108 NBAG03000258 BAK62695.1 JAA07056.1 JAA20456.1 JAA28770.1 JAA37631.1 PNI58053.1
KQ460878 KPJ11567.1 ODYU01004448 SOQ44328.1 RSAL01000115 RVE46897.1 PCG76977.1 GEZM01040026 JAV80992.1 KQ971348 EFA04843.1 KK852622 KDR19990.1 APGK01052855 KB741216 ENN72479.1 NEVH01022362 PNF18902.1 KB632404 ERL95062.1 GAPW01001224 JAC12374.1 PYGN01000201 PSN51347.1 CH477464 EAT40513.1 GDUN01001104 JAN94815.1 GALA01001171 JAA93681.1 PNF18899.1 GEZM01040028 JAV80990.1 DS235321 EEB14655.1 GFJQ02000088 JAW06882.1 ABLF02038839 GEFH01004042 JAP64539.1 GGMS01011999 MBY81202.1 GFDF01003875 JAV10209.1 GGLE01005552 MBY09678.1 GGMR01004376 MBY16995.1 GACK01001044 JAA63990.1 CVRI01000030 CRK92509.1 CRK92508.1 GBYB01004572 GBYB01012001 JAG74339.1 JAG81768.1 GECU01036271 JAS71435.1 BT127848 AEE62810.1 GFWV01009147 MAA33876.1 JW863603 AFO96120.1 GFPF01013267 MAA24413.1 GEDV01003625 JAP84932.1 AB047550 AB076609 AK043091 AK077443 AK159468 AL592222 AL604063 BC004013 AGTP01022571 AGTP01022572 AGTP01022573 AGTP01022574 AGTP01022575 AGTP01022576 AGTP01022577 AGTP01022578 AGTP01022579 AGTP01022580 ABJB010145156 ABJB010187107 ABJB010333299 ABJB010368120 ABJB010697092 DS785766 EEC09888.1 AAMC01027125 AAMC01027126 AAMC01027127 AAMC01027128 AAMC01027129 AAMC01027130 AAMC01027131 AAMC01027132 BC080142 GFAA01001026 JAU02409.1 GECZ01021818 JAS47951.1 AYCK01007209 AYCK01007210 AYCK01007211 AYCK01007212 AYCK01007213 GDIP01039654 JAM64061.1 AHAT01005716 AHAT01005717 CM004469 OCT92226.1 KK121348 KFM80280.1 GDIQ01078297 JAN16440.1 MWRG01014306 PRD24687.1 GDIP01223326 JAJ00076.1 GDIP01205824 GDIP01143042 LRGB01003071 JAJ17578.1 KZS04754.1 BEZZ01000472 GCC32917.1 GDIP01069753 JAM33962.1 GDIQ01159617 JAK92108.1 AF411216 BC061721 GDIQ01248190 JAK03535.1 KN122207 KFO32167.1 GDIP01139788 JAL63926.1 GDIQ01203662 JAK48063.1 CP012528 ALC49071.1 GDIQ01184271 JAK67454.1 BFAA01001385 GCB64631.1 KE161298 EPQ02932.1 ADFV01107744 ADFV01107745 ADFV01107746 ADFV01107747 ADFV01107748 ADFV01107749 ADFV01107750 ADFV01107751 ADFV01107752 ADFV01107753 NDHI03003397 PNJ65719.1 CR859383 GAMS01007511 GAMR01000421 GAMQ01003879 JAB15625.1 JAB33511.1 JAB37972.1 AGCU01199602 AGCU01199603 AGCU01199604 AGCU01199605 CABD030036879 CABD030036880 CABD030036881 CABD030036882 CABD030036883 CABD030036884 CABD030036885 CABD030036886 AJFE02058748 AJFE02058749 AJFE02058750 AJFE02058751 AJFE02058752 AJFE02058753 AJFE02058754 AJFE02058755 AJFE02058756 AJFE02058757 AK305701 GABC01004282 GABF01001689 GABD01004330 GABE01007108 NBAG03000258 BAK62695.1 JAA07056.1 JAA20456.1 JAA28770.1 JAA37631.1 PNI58053.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000053240
UP000283053
UP000192223
+ More
UP000007266 UP000027135 UP000019118 UP000235965 UP000030742 UP000245037 UP000008820 UP000009046 UP000007819 UP000183832 UP000261480 UP000242638 UP000002852 UP000261500 UP000000589 UP000095300 UP000248484 UP000245320 UP000005215 UP000001555 UP000008143 UP000248480 UP000028760 UP000018468 UP000186698 UP000054359 UP000252040 UP000248483 UP000076858 UP000287033 UP000265300 UP000002494 UP000245341 UP000265020 UP000245340 UP000286641 UP000028990 UP000092553 UP000248481 UP000288216 UP000189704 UP000233040 UP000233220 UP000001073 UP000007646 UP000001595 UP000008225 UP000007267 UP000095301 UP000079721 UP000001519 UP000240080
UP000007266 UP000027135 UP000019118 UP000235965 UP000030742 UP000245037 UP000008820 UP000009046 UP000007819 UP000183832 UP000261480 UP000242638 UP000002852 UP000261500 UP000000589 UP000095300 UP000248484 UP000245320 UP000005215 UP000001555 UP000008143 UP000248480 UP000028760 UP000018468 UP000186698 UP000054359 UP000252040 UP000248483 UP000076858 UP000287033 UP000265300 UP000002494 UP000245341 UP000265020 UP000245340 UP000286641 UP000028990 UP000092553 UP000248481 UP000288216 UP000189704 UP000233040 UP000233220 UP000001073 UP000007646 UP000001595 UP000008225 UP000007267 UP000095301 UP000079721 UP000001519 UP000240080
SUPFAM
SSF68906
SSF68906
Gene 3D
ProteinModelPortal
A0A2A4JZY8
A0A212FBQ9
A0A194Q645
A0A194R1K1
A0A2H1VU66
A0A437B8N5
+ More
A0A2A4JY81 A0A1W4WRU7 A0A1Y1M5D5 D2A456 A0A067R8P9 N6SXI5 A0A1W4X2T3 A0A2J7PRE6 U4UPM4 A0A023EV67 A0A2P8Z4E6 Q0IEV8 A0A0P6IY74 T1DI36 A0A2J7PRF6 A0A1Y1M8U0 E0VMP9 A0A1Z5LHI1 J9K896 A0A131XC51 A0A2S2QVQ3 A0A1L8DUW3 A0A2R5LJT0 A0A2S2NIL2 L7MJ41 A0A1J1HWW3 A0A1J1HYV6 A0A0C9QV73 A0A1B6H9L4 J3JX80 A0A293M0N7 A0A3B3YLG3 V9KEE5 A0A3P9NVI1 A0A224ZB22 M3ZD62 A0A3B3ULI2 A0A131Z2A8 Q99KU0 A0A1I8P765 A0A2Y9T4G7 A0A2U4C5Y9 I3MR59 B7PTG6 F6PQC8 Q68EQ9 A0A1E1XSX9 A0A1B6FCN4 A0A2Y9T6B8 A0A2Y9DPJ9 A0A087Y456 A0A0P5ZT58 W5M8B4 A0A1L8H860 A0A087USE1 A0A0P6DET1 A0A2P6KEG0 A0A341CFG8 A0A2Y9LFD4 A0A0P4YUV6 A0A0P5A927 A0A401SRA5 A0A0P5XN04 A0A0P5M7J0 A0A340WK98 Q91ZQ0 A0A2U3Y9I3 A0A3Q2FW45 A0A2U3VRG2 A0A0P5HMK8 A0A3Q7MK72 A0A091DM61 A0A0P5SH55 A0A0P5J8B9 A0A0M4EMP9 A0A2Y9IBG3 A0A0P5KUU4 A0A401NUR4 A0A1U7UT36 S7P5P1 A0A2K5QG81 A0A2K6STR0 A0A2I3HZ10 G3TCW1 A0A2J8W7G8 Q5R9K4 F7EZ12 K7FP29 A0A1I8MIE4 A0A1S3AEJ0 G3QIS0 A0A2R9APT4 G2HG37
A0A2A4JY81 A0A1W4WRU7 A0A1Y1M5D5 D2A456 A0A067R8P9 N6SXI5 A0A1W4X2T3 A0A2J7PRE6 U4UPM4 A0A023EV67 A0A2P8Z4E6 Q0IEV8 A0A0P6IY74 T1DI36 A0A2J7PRF6 A0A1Y1M8U0 E0VMP9 A0A1Z5LHI1 J9K896 A0A131XC51 A0A2S2QVQ3 A0A1L8DUW3 A0A2R5LJT0 A0A2S2NIL2 L7MJ41 A0A1J1HWW3 A0A1J1HYV6 A0A0C9QV73 A0A1B6H9L4 J3JX80 A0A293M0N7 A0A3B3YLG3 V9KEE5 A0A3P9NVI1 A0A224ZB22 M3ZD62 A0A3B3ULI2 A0A131Z2A8 Q99KU0 A0A1I8P765 A0A2Y9T4G7 A0A2U4C5Y9 I3MR59 B7PTG6 F6PQC8 Q68EQ9 A0A1E1XSX9 A0A1B6FCN4 A0A2Y9T6B8 A0A2Y9DPJ9 A0A087Y456 A0A0P5ZT58 W5M8B4 A0A1L8H860 A0A087USE1 A0A0P6DET1 A0A2P6KEG0 A0A341CFG8 A0A2Y9LFD4 A0A0P4YUV6 A0A0P5A927 A0A401SRA5 A0A0P5XN04 A0A0P5M7J0 A0A340WK98 Q91ZQ0 A0A2U3Y9I3 A0A3Q2FW45 A0A2U3VRG2 A0A0P5HMK8 A0A3Q7MK72 A0A091DM61 A0A0P5SH55 A0A0P5J8B9 A0A0M4EMP9 A0A2Y9IBG3 A0A0P5KUU4 A0A401NUR4 A0A1U7UT36 S7P5P1 A0A2K5QG81 A0A2K6STR0 A0A2I3HZ10 G3TCW1 A0A2J8W7G8 Q5R9K4 F7EZ12 K7FP29 A0A1I8MIE4 A0A1S3AEJ0 G3QIS0 A0A2R9APT4 G2HG37
Ontologies
GO
Topology
Subcellular location
Endoplasmic reticulum-Golgi intermediate compartment membrane
Cell membrane
Vacuole membrane
Endoplasmic reticulum
Cell membrane
Vacuole membrane
Endoplasmic reticulum
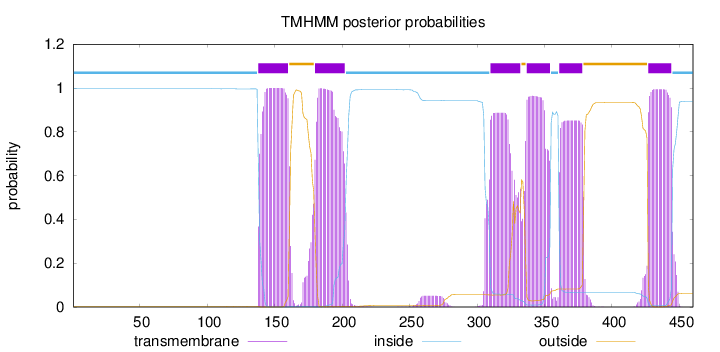
Length:
460
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
120.93962
Exp number, first 60 AAs:
0
Total prob of N-in:
0.99661
inside
1 - 137
TMhelix
138 - 160
outside
161 - 179
TMhelix
180 - 202
inside
203 - 309
TMhelix
310 - 332
outside
333 - 336
TMhelix
337 - 354
inside
355 - 360
TMhelix
361 - 378
outside
379 - 426
TMhelix
427 - 444
inside
445 - 460
Population Genetic Test Statistics
Pi
280.39883
Theta
186.489336
Tajima's D
1.832582
CLR
0.082359
CSRT
0.853207339633018
Interpretation
Uncertain
