Gene
KWMTBOMO09685
Pre Gene Modal
BGIBMGA012878
Annotation
K3_protein_[Bombyx_mori]
Full name
SH3 domain-containing protein Dlish
Alternative Name
Dachs ligand with SH3 domains
Location in the cell
Nuclear Reliability : 3.155
Sequence
CDS
ATGGCATTTCTATGTCCAGTCAGGATCAGAAGGGGAAAGAAGAAGAAATCAAGTTCTGGAGACTCTGACAAAGACCTCTCCAGGCCCAGTTCAGGACTGAGTCCTGGGATGGGCCGCATCACAGGATCAGCAAGCATAGAAACATTAGTGAGAGTTGGCATTGAGAAGGAACATGGCCTCAGTCCTGATTCGAAGATGGTGGTGCTCCATGATTTCACACCATGTGTGGATGACGAGTTAGAGGTAAAACGTGGACAAATTGTTAATGTTCTATATAGAGAAAATGATTGGGTTTATGTCATAGTAGCAGAATCTCGAAGAGAAGGCTTCATACCGCATTCATATTGTGCACCATGTGAACATCATGATTTAAAGAAAAAACTCCCGAGAAGTAGATCCCCTGCCGATTTAGCCCATCGAGATGTTTCACAATTATCTGTATCTGATGGAGTAACAAATGATGGGCATTCAGAGCTTGGTAGTGAAGGTGAAGCATGTCCCTTTAGCAAAGATCCTTCAGGAAGATACGTAGTGCTATACACATTCACTGCAAGAGATGAAAATGATGTTGACGTAGAACGAGGAGAATTTGTTACAGTACTAAACAGAGAAGACCCAGATTGGTATTGGATAGTAAGGAGTGATGGTCAAGAAGGGTTTATACCATCTGGTTTTGTGTATCCTGCTGTCGTTCAAGCCACCACACAAGAGAACACACAGACCTTGCATCCTACGCCGCCAACAAATACAAATAACAGTATAATATCAACAAATAACAACAGTAGTATAAACAACACAACACAGCAGGACAACGACGGGAGGTATCACGGAACGGAACTAGTCATGCTTTATGATTACAAGGCTCAAGCACCTGACGATCTGACAGTGAAGCGAGGAGAGTGGGTTTATGCAGATCTGACCCAGCAAACTGTAGAGGGATGGTTGTGGGCACACGCACCCAAGTCACGACGATCCGGTTTCATACCTACAGCTTACGCTCGTGCACCTCACACTACATCACTTTGA
Protein
MAFLCPVRIRRGKKKKSSSGDSDKDLSRPSSGLSPGMGRITGSASIETLVRVGIEKEHGLSPDSKMVVLHDFTPCVDDELEVKRGQIVNVLYRENDWVYVIVAESRREGFIPHSYCAPCEHHDLKKKLPRSRSPADLAHRDVSQLSVSDGVTNDGHSELGSEGEACPFSKDPSGRYVVLYTFTARDENDVDVERGEFVTVLNREDPDWYWIVRSDGQEGFIPSGFVYPAVVQATTQENTQTLHPTPPTNTNNSIISTNNNSSINNTTQQDNDGRYHGTELVMLYDYKAQAPDDLTVKRGEWVYADLTQQTVEGWLWAHAPKSRRSGFIPTAYARAPHTTSL
Summary
Description
Required for the apical cell cortex localization, total cellular level and full activity of dachs.
Subunit
Interacts with dachs (via C-terminus); the interaction is direct. Interacts (via N-terminus including SH3 domain 1) with palmitoyltransferase app; this leads to palmitoylation of Dlish by app. Also interacts with dco, ft, ft-regulated E3 ubiquitin ligase Fbxl7, F-box protein slmb and SCF E3 ubiquitin-protein ligase complex component Cul1.
Keywords
Complete proteome
Cytoplasm
Lipoprotein
Palmitate
Reference proteome
Repeat
SH3 domain
Feature
chain SH3 domain-containing protein Dlish
Uniprot
B0I191
A0A194PZX7
A0A437B952
A0A2H1VRL2
S4P982
A0A2A4JZA6
+ More
A0A212FBS2 H9JTL6 A0A194R2U4 A0A1Y1LW69 D6WMB9 A0A1Y1LW73 A0A1W4XET1 Q17LQ1 U4UN05 B0W5J6 A0A0N8ERZ4 A0A1L8DMH2 A0A2P8XSB3 A0A0J9RFI6 A1ZAY1 A0A182G0H6 Q8IGN9 B3NMM1 A0A1B0CSQ1 B4HMZ4 B4P551 A0A023F5F9 A0A2J7R2B5 A0A0A9XDR3 A0A1B6IIN7 A0A1W4V0T3 A0A067RIB4 A0A1S4G2Z6 B3MIE0 U5ETZ4 A0A1B6DKP4 B4GIA2 A0A1A9WK47 B5DZC0 A0A3S4R4W4 A0A3B0JQF8 A0A443SBK2 B4J5M3 B4K1U4 A0A2H8TZY2 J9JRL0 A0A0L0CK89 A0A224YZS3 A0A131YSH8 A0A1I8Q9Q2 L7M7S1 A0A2R5LL15 A0A1A9ZP91 B4NN52 B4LP50 T1JHQ5 A0A131XT93 A0A1J1J7X0 A0A131XI31 A0A1A9VX67 A0A1J1JAE9 A0A1B0GDL2 A0A2P2I2W9 A0A087TDS4 A0A1B0BR38 A0A1A9Y6C5 A0A182MM71 K7IZ02 A0A232FJ63 A0A444T7E9 A0A0T6B8U4 A0A0J7NM32 A0A310SA05 A0A195BP98 F4WL62 A0A026VST7 A0A151WLW5 A0A195CI93 A0A158P094 E9IKV0 E2BWK2 A0A088AFW4 A0A2A3EGM0 A0A154P8C3 E2AR06 A0A195FCU5 A0A1B0DHT7 A0A3L8D754 Q16FB7 A0A3Q0J412 T1H5W8 A0A0K8TSR1 Q7PNR5 A0A2M3ZBS8 A0A195EK69 A0A2S2NQ88 W5JFJ3 A0A2M4AF42 A0A182WB21
A0A212FBS2 H9JTL6 A0A194R2U4 A0A1Y1LW69 D6WMB9 A0A1Y1LW73 A0A1W4XET1 Q17LQ1 U4UN05 B0W5J6 A0A0N8ERZ4 A0A1L8DMH2 A0A2P8XSB3 A0A0J9RFI6 A1ZAY1 A0A182G0H6 Q8IGN9 B3NMM1 A0A1B0CSQ1 B4HMZ4 B4P551 A0A023F5F9 A0A2J7R2B5 A0A0A9XDR3 A0A1B6IIN7 A0A1W4V0T3 A0A067RIB4 A0A1S4G2Z6 B3MIE0 U5ETZ4 A0A1B6DKP4 B4GIA2 A0A1A9WK47 B5DZC0 A0A3S4R4W4 A0A3B0JQF8 A0A443SBK2 B4J5M3 B4K1U4 A0A2H8TZY2 J9JRL0 A0A0L0CK89 A0A224YZS3 A0A131YSH8 A0A1I8Q9Q2 L7M7S1 A0A2R5LL15 A0A1A9ZP91 B4NN52 B4LP50 T1JHQ5 A0A131XT93 A0A1J1J7X0 A0A131XI31 A0A1A9VX67 A0A1J1JAE9 A0A1B0GDL2 A0A2P2I2W9 A0A087TDS4 A0A1B0BR38 A0A1A9Y6C5 A0A182MM71 K7IZ02 A0A232FJ63 A0A444T7E9 A0A0T6B8U4 A0A0J7NM32 A0A310SA05 A0A195BP98 F4WL62 A0A026VST7 A0A151WLW5 A0A195CI93 A0A158P094 E9IKV0 E2BWK2 A0A088AFW4 A0A2A3EGM0 A0A154P8C3 E2AR06 A0A195FCU5 A0A1B0DHT7 A0A3L8D754 Q16FB7 A0A3Q0J412 T1H5W8 A0A0K8TSR1 Q7PNR5 A0A2M3ZBS8 A0A195EK69 A0A2S2NQ88 W5JFJ3 A0A2M4AF42 A0A182WB21
Pubmed
26354079
23622113
22118469
19121390
28004739
18362917
+ More
19820115 17510324 23537049 26999592 29403074 22936249 10731132 12537572 27692068 27824307 26483478 17994087 17550304 25474469 25401762 26823975 24845553 15632085 26108605 28797301 26830274 25576852 28049606 20075255 28648823 21719571 24508170 21347285 21282665 20798317 30249741 26369729 12364791 20920257 23761445
19820115 17510324 23537049 26999592 29403074 22936249 10731132 12537572 27692068 27824307 26483478 17994087 17550304 25474469 25401762 26823975 24845553 15632085 26108605 28797301 26830274 25576852 28049606 20075255 28648823 21719571 24508170 21347285 21282665 20798317 30249741 26369729 12364791 20920257 23761445
EMBL
AB162719
BAG06921.1
KQ459582
KPI98882.1
RSAL01000115
RVE46895.1
+ More
ODYU01004004 SOQ43438.1 GAIX01009190 JAA83370.1 NWSH01000362 PCG76974.1 AGBW02009293 OWR51168.1 BABH01003616 BABH01003617 KQ460878 KPJ11565.1 GEZM01045472 JAV77794.1 KQ971343 EFA04249.1 GEZM01045473 JAV77793.1 CH477213 EAT47651.1 KB632288 ERL91551.1 DS231843 EDS35479.1 GDUN01000898 JAN95021.1 GFDF01006425 JAV07659.1 PYGN01001431 PSN34887.1 CM002911 KMY94661.1 AE013599 BT099964 ACX47665.1 JXUM01001336 JXUM01001337 JXUM01001338 JXUM01001339 BT001682 AAN71437.1 CH954179 EDV54960.1 AJWK01026374 AJWK01026375 AJWK01026376 AJWK01026377 AJWK01026378 CH480816 EDW48344.1 CM000158 EDW91752.1 GBBI01002334 JAC16378.1 NEVH01007841 PNF34961.1 GBHO01028377 GBHO01028374 GBRD01010169 GDHC01011508 GDHC01008950 JAG15227.1 JAG15230.1 JAG55655.1 JAQ07121.1 JAQ09679.1 GECU01020917 JAS86789.1 KK852665 KDR19006.1 CH902619 EDV36988.1 GANO01001700 JAB58171.1 GEDC01011039 JAS26259.1 CH479183 EDW36222.1 CM000071 EDY69167.1 NCKU01001905 NCKU01001883 NCKU01001620 RWS10946.1 RWS11011.1 RWS11645.1 OUUW01000001 SPP74891.1 NCKV01004237 RWS24941.1 CH916367 EDW01799.1 CH917862 EDW04895.1 GFXV01007506 MBW19311.1 ABLF02035819 JRES01000265 KNC32818.1 GFPF01008166 MAA19312.1 GEDV01007009 JAP81548.1 GACK01005885 JAA59149.1 GGLE01006053 MBY10179.1 CH964282 EDW85791.1 CH940648 EDW62244.1 JH431388 GEFM01007005 JAP68791.1 CVRI01000075 CRL08533.1 GEFH01002821 JAP65760.1 CRL08534.1 CCAG010011939 IACF01002704 LAB68349.1 KK114767 KFM63263.1 JXJN01018905 AXCM01000549 NNAY01000120 OXU30784.1 SAUD01009372 RXG61563.1 LJIG01009135 KRT83685.1 LBMM01003433 KMQ93545.1 KQ767320 OAD53401.1 KQ976433 KYM87279.1 GL888207 EGI65025.1 KK108267 EZA46792.1 KQ982944 KYQ48882.1 KQ977720 KYN00460.1 ADTU01004650 ADTU01004651 GL764026 EFZ18861.1 GL451131 EFN79958.1 KZ288254 PBC30860.1 KQ434844 KZC08175.1 GL441846 EFN64152.1 KQ981685 KYN38022.1 AJVK01061554 QOIP01000012 RLU16104.1 CH478467 EAT32926.1 GDAI01000623 JAI16980.1 AAAB01008960 EAA11721.5 GGFM01005174 MBW25925.1 KQ978747 KYN28660.1 GGMR01006731 MBY19350.1 ADMH02001639 ETN61650.1 GGFK01006074 MBW39395.1
ODYU01004004 SOQ43438.1 GAIX01009190 JAA83370.1 NWSH01000362 PCG76974.1 AGBW02009293 OWR51168.1 BABH01003616 BABH01003617 KQ460878 KPJ11565.1 GEZM01045472 JAV77794.1 KQ971343 EFA04249.1 GEZM01045473 JAV77793.1 CH477213 EAT47651.1 KB632288 ERL91551.1 DS231843 EDS35479.1 GDUN01000898 JAN95021.1 GFDF01006425 JAV07659.1 PYGN01001431 PSN34887.1 CM002911 KMY94661.1 AE013599 BT099964 ACX47665.1 JXUM01001336 JXUM01001337 JXUM01001338 JXUM01001339 BT001682 AAN71437.1 CH954179 EDV54960.1 AJWK01026374 AJWK01026375 AJWK01026376 AJWK01026377 AJWK01026378 CH480816 EDW48344.1 CM000158 EDW91752.1 GBBI01002334 JAC16378.1 NEVH01007841 PNF34961.1 GBHO01028377 GBHO01028374 GBRD01010169 GDHC01011508 GDHC01008950 JAG15227.1 JAG15230.1 JAG55655.1 JAQ07121.1 JAQ09679.1 GECU01020917 JAS86789.1 KK852665 KDR19006.1 CH902619 EDV36988.1 GANO01001700 JAB58171.1 GEDC01011039 JAS26259.1 CH479183 EDW36222.1 CM000071 EDY69167.1 NCKU01001905 NCKU01001883 NCKU01001620 RWS10946.1 RWS11011.1 RWS11645.1 OUUW01000001 SPP74891.1 NCKV01004237 RWS24941.1 CH916367 EDW01799.1 CH917862 EDW04895.1 GFXV01007506 MBW19311.1 ABLF02035819 JRES01000265 KNC32818.1 GFPF01008166 MAA19312.1 GEDV01007009 JAP81548.1 GACK01005885 JAA59149.1 GGLE01006053 MBY10179.1 CH964282 EDW85791.1 CH940648 EDW62244.1 JH431388 GEFM01007005 JAP68791.1 CVRI01000075 CRL08533.1 GEFH01002821 JAP65760.1 CRL08534.1 CCAG010011939 IACF01002704 LAB68349.1 KK114767 KFM63263.1 JXJN01018905 AXCM01000549 NNAY01000120 OXU30784.1 SAUD01009372 RXG61563.1 LJIG01009135 KRT83685.1 LBMM01003433 KMQ93545.1 KQ767320 OAD53401.1 KQ976433 KYM87279.1 GL888207 EGI65025.1 KK108267 EZA46792.1 KQ982944 KYQ48882.1 KQ977720 KYN00460.1 ADTU01004650 ADTU01004651 GL764026 EFZ18861.1 GL451131 EFN79958.1 KZ288254 PBC30860.1 KQ434844 KZC08175.1 GL441846 EFN64152.1 KQ981685 KYN38022.1 AJVK01061554 QOIP01000012 RLU16104.1 CH478467 EAT32926.1 GDAI01000623 JAI16980.1 AAAB01008960 EAA11721.5 GGFM01005174 MBW25925.1 KQ978747 KYN28660.1 GGMR01006731 MBY19350.1 ADMH02001639 ETN61650.1 GGFK01006074 MBW39395.1
Proteomes
UP000053268
UP000283053
UP000218220
UP000007151
UP000005204
UP000053240
+ More
UP000007266 UP000192223 UP000008820 UP000030742 UP000002320 UP000245037 UP000000803 UP000069940 UP000008711 UP000092461 UP000001292 UP000002282 UP000235965 UP000192221 UP000027135 UP000007801 UP000008744 UP000091820 UP000001819 UP000285301 UP000268350 UP000288716 UP000001070 UP000007819 UP000037069 UP000095300 UP000092445 UP000007798 UP000008792 UP000183832 UP000078200 UP000092444 UP000054359 UP000092460 UP000092443 UP000075883 UP000002358 UP000215335 UP000288706 UP000036403 UP000078540 UP000007755 UP000053097 UP000075809 UP000078542 UP000005205 UP000008237 UP000005203 UP000242457 UP000076502 UP000000311 UP000078541 UP000092462 UP000279307 UP000079169 UP000015102 UP000007062 UP000078492 UP000000673 UP000075920
UP000007266 UP000192223 UP000008820 UP000030742 UP000002320 UP000245037 UP000000803 UP000069940 UP000008711 UP000092461 UP000001292 UP000002282 UP000235965 UP000192221 UP000027135 UP000007801 UP000008744 UP000091820 UP000001819 UP000285301 UP000268350 UP000288716 UP000001070 UP000007819 UP000037069 UP000095300 UP000092445 UP000007798 UP000008792 UP000183832 UP000078200 UP000092444 UP000054359 UP000092460 UP000092443 UP000075883 UP000002358 UP000215335 UP000288706 UP000036403 UP000078540 UP000007755 UP000053097 UP000075809 UP000078542 UP000005205 UP000008237 UP000005203 UP000242457 UP000076502 UP000000311 UP000078541 UP000092462 UP000279307 UP000079169 UP000015102 UP000007062 UP000078492 UP000000673 UP000075920
Interpro
Gene 3D
ProteinModelPortal
B0I191
A0A194PZX7
A0A437B952
A0A2H1VRL2
S4P982
A0A2A4JZA6
+ More
A0A212FBS2 H9JTL6 A0A194R2U4 A0A1Y1LW69 D6WMB9 A0A1Y1LW73 A0A1W4XET1 Q17LQ1 U4UN05 B0W5J6 A0A0N8ERZ4 A0A1L8DMH2 A0A2P8XSB3 A0A0J9RFI6 A1ZAY1 A0A182G0H6 Q8IGN9 B3NMM1 A0A1B0CSQ1 B4HMZ4 B4P551 A0A023F5F9 A0A2J7R2B5 A0A0A9XDR3 A0A1B6IIN7 A0A1W4V0T3 A0A067RIB4 A0A1S4G2Z6 B3MIE0 U5ETZ4 A0A1B6DKP4 B4GIA2 A0A1A9WK47 B5DZC0 A0A3S4R4W4 A0A3B0JQF8 A0A443SBK2 B4J5M3 B4K1U4 A0A2H8TZY2 J9JRL0 A0A0L0CK89 A0A224YZS3 A0A131YSH8 A0A1I8Q9Q2 L7M7S1 A0A2R5LL15 A0A1A9ZP91 B4NN52 B4LP50 T1JHQ5 A0A131XT93 A0A1J1J7X0 A0A131XI31 A0A1A9VX67 A0A1J1JAE9 A0A1B0GDL2 A0A2P2I2W9 A0A087TDS4 A0A1B0BR38 A0A1A9Y6C5 A0A182MM71 K7IZ02 A0A232FJ63 A0A444T7E9 A0A0T6B8U4 A0A0J7NM32 A0A310SA05 A0A195BP98 F4WL62 A0A026VST7 A0A151WLW5 A0A195CI93 A0A158P094 E9IKV0 E2BWK2 A0A088AFW4 A0A2A3EGM0 A0A154P8C3 E2AR06 A0A195FCU5 A0A1B0DHT7 A0A3L8D754 Q16FB7 A0A3Q0J412 T1H5W8 A0A0K8TSR1 Q7PNR5 A0A2M3ZBS8 A0A195EK69 A0A2S2NQ88 W5JFJ3 A0A2M4AF42 A0A182WB21
A0A212FBS2 H9JTL6 A0A194R2U4 A0A1Y1LW69 D6WMB9 A0A1Y1LW73 A0A1W4XET1 Q17LQ1 U4UN05 B0W5J6 A0A0N8ERZ4 A0A1L8DMH2 A0A2P8XSB3 A0A0J9RFI6 A1ZAY1 A0A182G0H6 Q8IGN9 B3NMM1 A0A1B0CSQ1 B4HMZ4 B4P551 A0A023F5F9 A0A2J7R2B5 A0A0A9XDR3 A0A1B6IIN7 A0A1W4V0T3 A0A067RIB4 A0A1S4G2Z6 B3MIE0 U5ETZ4 A0A1B6DKP4 B4GIA2 A0A1A9WK47 B5DZC0 A0A3S4R4W4 A0A3B0JQF8 A0A443SBK2 B4J5M3 B4K1U4 A0A2H8TZY2 J9JRL0 A0A0L0CK89 A0A224YZS3 A0A131YSH8 A0A1I8Q9Q2 L7M7S1 A0A2R5LL15 A0A1A9ZP91 B4NN52 B4LP50 T1JHQ5 A0A131XT93 A0A1J1J7X0 A0A131XI31 A0A1A9VX67 A0A1J1JAE9 A0A1B0GDL2 A0A2P2I2W9 A0A087TDS4 A0A1B0BR38 A0A1A9Y6C5 A0A182MM71 K7IZ02 A0A232FJ63 A0A444T7E9 A0A0T6B8U4 A0A0J7NM32 A0A310SA05 A0A195BP98 F4WL62 A0A026VST7 A0A151WLW5 A0A195CI93 A0A158P094 E9IKV0 E2BWK2 A0A088AFW4 A0A2A3EGM0 A0A154P8C3 E2AR06 A0A195FCU5 A0A1B0DHT7 A0A3L8D754 Q16FB7 A0A3Q0J412 T1H5W8 A0A0K8TSR1 Q7PNR5 A0A2M3ZBS8 A0A195EK69 A0A2S2NQ88 W5JFJ3 A0A2M4AF42 A0A182WB21
PDB
1GRI
E-value=4.23967e-08,
Score=137
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Low levels are found diffusely in the cytoplasm with high levels concentrated at the subapical cell cortex. With evidence from 3 publications.
Cell cortex Low levels are found diffusely in the cytoplasm with high levels concentrated at the subapical cell cortex. With evidence from 3 publications.
Cell cortex Low levels are found diffusely in the cytoplasm with high levels concentrated at the subapical cell cortex. With evidence from 3 publications.
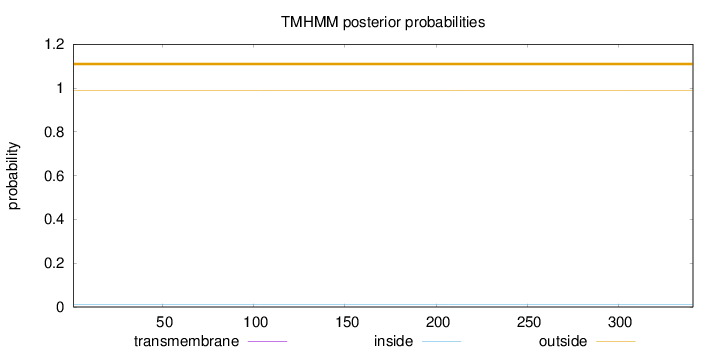
Length:
341
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00013
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01162
outside
1 - 341
Population Genetic Test Statistics
Pi
336.213716
Theta
228.844802
Tajima's D
1.546034
CLR
0.463797
CSRT
0.79846007699615
Interpretation
Uncertain
