Gene
KWMTBOMO09682
Annotation
PREDICTED:_uncharacterized_protein_LOC101745010_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.874 PlasmaMembrane Reliability : 2.23
Sequence
CDS
ATGCCAGATTCACTGTGCGCATGCGCATTGGAACAGCAGTGTTTTAAGACGGTATGTAATCAGCAGCCCACTAAAACTATATTGCTTGGTACAGCGATATGCAATATTATGGACAATTCAGGCACGTTCTTGAGTACTGCCCGAATATTGGTAGATAGTGCATCGCAACGAGATCTGATTACTCTTGAATTTTGTAAGAGACTTAATTTACCAATTTATCCTTCCGAAACAAAGCAAGTGTGCGGAGTGGGTGATGTGAAAAATTCCATTGAAGGATATTGTTGTCTAACACTTTGCTCTTGTACTGAAGAATCAGTGCGTATTACAATACAACCTCTGGTAATCAATAAAATTACAGGTGAACTTCCTACCTCAAAGATCGATACTTCAAGTCTTAATTATCTAAAACACATTAAGCTAGCTGATGACAGATTCTCACAGCCTAGTGGTATAGATTTGATTGTGGGTTCCGATGTATTCTCTCGAATTTTACGACCTCATATAATTTCTAGGGCCCCCGGTGAACCTGTAGCAATTGAGACTTCATTAGGTTACATAATAGTCGGCCAGGCACCGGTATTAGCTCCCTCGATAGATAATTCATACTATACACATTGCACCTTTGGTGAGGACAATTTGGATAATGAGAATAATGCTTGTATTGGTAATTTTTTGAAATTAGAGGATATTCCAGACGTCAAGCCATACACGGAACAGGAAGAGGAGTGTGAAAGCCTTTATAGACAAACGACTACTCGTGATAGCTTAGGTCGTTACATTGTCTCGTTACCATTCAAAGATACTCCTAGTAAGCTGGGTGACTCATTACAGGCATCAATGCGTAGATATCTTTCATTGGAACGTAAGCTGCTATTGCAACCCGAAATGAGACAAGAATACGATAAAGTCATTAAGGACTATTTAGAAAAGGGATATTTAAACCCATGTTCTAGAAATATAAATGAGTCCTCACTCCACTATGTTATTCCCCATCACGGCATCGTTCGAAAGGATAAAAGTACAACAAAATTACGGGTAGTATTGGATGGCAGTATGAAATCATCATCGGGACTGGCACTGAATGATATATTACAAGTCGGACCGAATTTGCAAAACGACCTTTTTAGAATAATTTTAAATTTCAGACTTTTTAATGTAGCGATTAGTGCCGACATTAGGCAAATGTACTTGAGGATTTTAGTAAAGGACGAAGATAGGAAATTCCTGAGAATGTTGTATCGCTTTGATCCAAGCGAGGAGATAAAACTTTACGAATTCACACGTGTGCCATTCGGTTTGTGTTGTAGTCCGTTCCTTGCGATCAGGACTGTGCGCCAGTTAGTGACGGACGAGGGATCACAATTTCCTCTCGCGGCTCCAGTAGCTGACAGTGACGTTTTCACTGACGATTTGGCTACTTCCTGTGCGAATGAAGAAACAGCGGTCGAACTTTCTAATCAACTTATTAAAATGTTTCATGCTGGTGGTTTTGATTTGGTAAAGTTTTCAAGTAACTCACCAGAGGTTCTTTCTAAGATACCCTCTTCACATAGAGAATTCGAAGTTATAGAATTTAGTCCAGATAGCTATTTAAAAATACTGGGACTAAACTGGCTTCCAGCCGAAGACGTATTCACATTCACAGTAAATTTACGGAACCGCGAATGTACTAAAAGAAATATTTTATCAGTTATAGCAAGAATATGGGACTTAATGGGATTCGTAGCTCCTGTTACACTATGGGCCAAGCTCATCATTAAGTCACTATGGGCTAATAATATAGATTGGGATGAAACTCCTCGCCCTGAAATTGTAGCAGCTTGGCATCGATTCGTGTCTGAGTTGCCAATATTGGAAAATGTTAGAATACCACGACATACTGGCATTGTTATAGAGTGCATAGTTAGCTTATTGGGATTTGCTGATGCATCTGAAAAGGCCTATGGAGGTGTCGTTTATTTACATGTTTATTTTCCACAAACAAATAAGTTCACTATTACGCTTGTTTGTGCCAAATCCAGAGTAGCGCCGTTACGATGCGTGTCATTGGCAAGACTAGAATTATGCGGTAACTTGATTCTCGCAAAGTTGATGAGAGCAATTATTGACAGCTATTCTCATCGCTGTAAAATAAGTAATGTATTTGCCTTTACGGATAGCACCGTGGCTTTGGCATGGATTCACTCATCACCGGCTAGATGGCATACGTTTGTAGCCAACCGAGTAACAAAGATACAGGATGAAATACACCCCAGAAACTTTTATTTTGTATCGGGTAAAGAGAATCCAGCAGACTGTCTTTCACGCGGCTTAACACCATCACAATTGATGGAGCATCCTCTTTGGTTCAGTGGACCCCGATTTGCTCATCTACCCATTTCAGATTGGCCAGTTAAGAATTTTGATGCCACATCGATGACTGATATTCCAGAAATGAAACCAGCTATCATGCTATTCACAACTGATGATGCACGGAATGACAATATATTATACACGCTTGCATGCCGTATTTCATCATGGCCCAAGCTGGTCCGTATTGTACTTTATGTTCTCCGGTTTATTAAATTGGTACCGAGTAAATTTGATATTTCCCATTTAAAGGTGGCCGAAACGAATATCATCAAGGCTGTACAGCGAGTTCACTTTGATAAAGAACTTAAGAATGTGAAGTCTGGCAAAATATTACCGTCTGCATTTCAACGGCTCAGAGCCTTTGTTCACGAAGAAACGATAACGGAACATTCTAATCTCATCCAAAGGAAACGTATTGTAGACAGTTTGGTCCAGTCTTTTTGGAAAAGATGGAAACTTGAATATCTTAATACATTACAAGTTCGTAATAAATGGAAAAAAGATAGCACTCCTATTAAAACTGGCACAGTTGTACTGCTTCAATCTGAAAATAGTGCTCCTCTTAATTGGCCATTGGGTATAATAGAGGAAACTTTTCCGGGTCGTGATGGAGTTGTAAGGGTGGTTAACGTTAGAACGAAAACTGGTACTTATCGTAGACCTGTTGTTAAGGTTTATCCTTTACCAACACAGTAG
Protein
MPDSLCACALEQQCFKTVCNQQPTKTILLGTAICNIMDNSGTFLSTARILVDSASQRDLITLEFCKRLNLPIYPSETKQVCGVGDVKNSIEGYCCLTLCSCTEESVRITIQPLVINKITGELPTSKIDTSSLNYLKHIKLADDRFSQPSGIDLIVGSDVFSRILRPHIISRAPGEPVAIETSLGYIIVGQAPVLAPSIDNSYYTHCTFGEDNLDNENNACIGNFLKLEDIPDVKPYTEQEEECESLYRQTTTRDSLGRYIVSLPFKDTPSKLGDSLQASMRRYLSLERKLLLQPEMRQEYDKVIKDYLEKGYLNPCSRNINESSLHYVIPHHGIVRKDKSTTKLRVVLDGSMKSSSGLALNDILQVGPNLQNDLFRIILNFRLFNVAISADIRQMYLRILVKDEDRKFLRMLYRFDPSEEIKLYEFTRVPFGLCCSPFLAIRTVRQLVTDEGSQFPLAAPVADSDVFTDDLATSCANEETAVELSNQLIKMFHAGGFDLVKFSSNSPEVLSKIPSSHREFEVIEFSPDSYLKILGLNWLPAEDVFTFTVNLRNRECTKRNILSVIARIWDLMGFVAPVTLWAKLIIKSLWANNIDWDETPRPEIVAAWHRFVSELPILENVRIPRHTGIVIECIVSLLGFADASEKAYGGVVYLHVYFPQTNKFTITLVCAKSRVAPLRCVSLARLELCGNLILAKLMRAIIDSYSHRCKISNVFAFTDSTVALAWIHSSPARWHTFVANRVTKIQDEIHPRNFYFVSGKENPADCLSRGLTPSQLMEHPLWFSGPRFAHLPISDWPVKNFDATSMTDIPEMKPAIMLFTTDDARNDNILYTLACRISSWPKLVRIVLYVLRFIKLVPSKFDISHLKVAETNIIKAVQRVHFDKELKNVKSGKILPSAFQRLRAFVHEETITEHSNLIQRKRIVDSLVQSFWKRWKLEYLNTLQVRNKWKKDSTPIKTGTVVLLQSENSAPLNWPLGIIEETFPGRDGVVRVVNVRTKTGTYRRPVVKVYPLPTQ
Summary
Similarity
Belongs to the synaptobrevin family.
Belongs to the G-protein coupled receptor 1 family.
Belongs to the G-protein coupled receptor 1 family.
Uniprot
A0A1S4EIX5
A0A1S3DJJ0
A0A1S3DJ34
A0A1S3DJC0
A0A1S3DIZ2
A0A023F096
+ More
A0A1S3DMK8 A0A1Y1LIC8 A0A1S3DMT1 A0A3Q0JCL2 A0A1S3DH93 A0A1S3DHM0 A0A3Q0J7N1 A0A3Q0J7K9 A0A3Q0JJJ7 A0A3Q0JFQ3 A0A3Q0J0P0 A0A1B6LLT3 A0A1B6MT91 A0A1B6KIX6 A0A1Y1LM33 A0A1Y1LIG5 A0A3Q0JBZ9 A0A437AV42 V5GHM7 A0A1Y1KMX0 X1WRA3 A0A1B6KZV2 A0A1B6MUZ9 X1WY95 A0A0J7K7U0 V5GC07 A0A2S2NCY5 J9JVD6 J9KD60 A0A2S2PM34 A0A0J7KDN6 X1XU26 X1XTX7 A0A0A9YYJ5 A0A2S2QPS5 A0A226CWA5 A0A087T9T6 A0A0J7KCR4 A0A1S3DGE3 J9JUK4 A0A2S2PNC2 J9JLG6 W8C1U1 J9K351 A0A224XG39 A0A226E226 A0A1B6GGX5 A0A023F0J1 A0A226D417 A0A226DMG9 A0A3Q0JI79 A0A1S3DHZ9 A0A1S3DIE2 A0A226CV54 A0A1S3DRF1 A0A3Q0JDB5 A0A146M0N4 X1WNM7 A0A0A9Z275 X1WY64 X1WZY4 A0A182MIR3 A0A1S3DM17 J9M7R2 A0A1U8N859 A0A0A9W8L8 A0A224XGH3 A0A226D0T3 A0A3Q0JP94 A0A2S2PLU2 A0A2S2P1P0 A0A226E0X4 A0A3L8DF49 A0A2S2PNZ4 A0A226EPI5 A0A146LBU6 A0A0N0PAG9 A0A0J7N465 A0A0A9VYL4 A0A146L5E8 A0A146L7F7 A0A0A9VSF7 A0A0A9X8U7 J9LPA9 A0A226EUK6 A0A226EM86 V5GU69
A0A1S3DMK8 A0A1Y1LIC8 A0A1S3DMT1 A0A3Q0JCL2 A0A1S3DH93 A0A1S3DHM0 A0A3Q0J7N1 A0A3Q0J7K9 A0A3Q0JJJ7 A0A3Q0JFQ3 A0A3Q0J0P0 A0A1B6LLT3 A0A1B6MT91 A0A1B6KIX6 A0A1Y1LM33 A0A1Y1LIG5 A0A3Q0JBZ9 A0A437AV42 V5GHM7 A0A1Y1KMX0 X1WRA3 A0A1B6KZV2 A0A1B6MUZ9 X1WY95 A0A0J7K7U0 V5GC07 A0A2S2NCY5 J9JVD6 J9KD60 A0A2S2PM34 A0A0J7KDN6 X1XU26 X1XTX7 A0A0A9YYJ5 A0A2S2QPS5 A0A226CWA5 A0A087T9T6 A0A0J7KCR4 A0A1S3DGE3 J9JUK4 A0A2S2PNC2 J9JLG6 W8C1U1 J9K351 A0A224XG39 A0A226E226 A0A1B6GGX5 A0A023F0J1 A0A226D417 A0A226DMG9 A0A3Q0JI79 A0A1S3DHZ9 A0A1S3DIE2 A0A226CV54 A0A1S3DRF1 A0A3Q0JDB5 A0A146M0N4 X1WNM7 A0A0A9Z275 X1WY64 X1WZY4 A0A182MIR3 A0A1S3DM17 J9M7R2 A0A1U8N859 A0A0A9W8L8 A0A224XGH3 A0A226D0T3 A0A3Q0JP94 A0A2S2PLU2 A0A2S2P1P0 A0A226E0X4 A0A3L8DF49 A0A2S2PNZ4 A0A226EPI5 A0A146LBU6 A0A0N0PAG9 A0A0J7N465 A0A0A9VYL4 A0A146L5E8 A0A146L7F7 A0A0A9VSF7 A0A0A9X8U7 J9LPA9 A0A226EUK6 A0A226EM86 V5GU69
EMBL
GBBI01004276
JAC14436.1
GEZM01054789
JAV73399.1
GEBQ01015458
JAT24519.1
+ More
GEBQ01000825 JAT39152.1 GEBQ01028569 JAT11408.1 GEZM01054766 GEZM01054765 JAV73410.1 JAV73413.1 RSAL01000392 RVE41994.1 GALX01004947 JAB63519.1 GEZM01085145 GEZM01085143 JAV60187.1 ABLF02066034 GEBQ01022991 GEBQ01006185 JAT16986.1 JAT33792.1 GEBQ01007199 GEBQ01000222 JAT32778.1 JAT39755.1 ABLF02030854 ABLF02067347 LBMM01012238 KMQ86334.1 GALX01000776 JAB67690.1 GGMR01002421 MBY15040.1 ABLF02008594 ABLF02041863 GGMR01017890 MBY30509.1 LBMM01009202 KMQ88311.1 ABLF02001506 ABLF02008029 ABLF02035899 ABLF02037719 ABLF02046327 ABLF02064627 ABLF02006304 ABLF02036268 GBHO01012499 GBHO01012498 GBHO01012497 GBHO01007479 GBHO01007451 GBHO01007450 JAG31105.1 JAG31106.1 JAG31107.1 JAG36125.1 JAG36153.1 JAG36154.1 GGMS01010524 MBY79727.1 LNIX01000069 OXA36894.1 KK114194 KFM61875.1 LBMM01009551 KMQ88039.1 ABLF02039606 GGMR01018294 MBY30913.1 ABLF02030698 ABLF02030705 ABLF02030709 GAMC01006864 JAB99691.1 ABLF02025193 GFTR01008874 JAW07552.1 LNIX01000007 OXA51795.1 GECZ01008071 JAS61698.1 GBBI01004211 JAC14501.1 LNIX01000036 OXA39969.1 LNIX01000016 OXA46198.1 LNIX01000074 OXA36769.1 GDHC01006469 JAQ12160.1 ABLF02042806 GBHO01007689 JAG35915.1 ABLF02012239 ABLF02012241 ABLF02017372 ABLF02042178 ABLF02066436 ABLF02036131 ABLF02041527 ABLF02064049 AXCM01013330 ABLF02005251 ABLF02055296 GBHO01039838 GBHO01006034 JAG03766.1 JAG37570.1 GFTR01008866 JAW07560.1 LNIX01000044 OXA38660.1 GGMR01017803 MBY30422.1 GGMR01010750 MBY23369.1 OXA51382.1 QOIP01000009 RLU19024.1 GGMR01018506 MBY31125.1 LNIX01000002 OXA59522.1 GDHC01014057 JAQ04572.1 KQ458473 KPJ05885.1 LBMM01010417 KMQ87475.1 GBHO01044316 JAF99287.1 GDHC01016272 JAQ02357.1 GDHC01014268 JAQ04361.1 GBHO01044317 GBHO01044315 JAF99286.1 JAF99288.1 GBHO01028371 GBHO01028370 JAG15233.1 JAG15234.1 ABLF02035780 LNIX01000001 OXA61293.1 LNIX01000003 OXA58579.1 GALX01003264 JAB65202.1
GEBQ01000825 JAT39152.1 GEBQ01028569 JAT11408.1 GEZM01054766 GEZM01054765 JAV73410.1 JAV73413.1 RSAL01000392 RVE41994.1 GALX01004947 JAB63519.1 GEZM01085145 GEZM01085143 JAV60187.1 ABLF02066034 GEBQ01022991 GEBQ01006185 JAT16986.1 JAT33792.1 GEBQ01007199 GEBQ01000222 JAT32778.1 JAT39755.1 ABLF02030854 ABLF02067347 LBMM01012238 KMQ86334.1 GALX01000776 JAB67690.1 GGMR01002421 MBY15040.1 ABLF02008594 ABLF02041863 GGMR01017890 MBY30509.1 LBMM01009202 KMQ88311.1 ABLF02001506 ABLF02008029 ABLF02035899 ABLF02037719 ABLF02046327 ABLF02064627 ABLF02006304 ABLF02036268 GBHO01012499 GBHO01012498 GBHO01012497 GBHO01007479 GBHO01007451 GBHO01007450 JAG31105.1 JAG31106.1 JAG31107.1 JAG36125.1 JAG36153.1 JAG36154.1 GGMS01010524 MBY79727.1 LNIX01000069 OXA36894.1 KK114194 KFM61875.1 LBMM01009551 KMQ88039.1 ABLF02039606 GGMR01018294 MBY30913.1 ABLF02030698 ABLF02030705 ABLF02030709 GAMC01006864 JAB99691.1 ABLF02025193 GFTR01008874 JAW07552.1 LNIX01000007 OXA51795.1 GECZ01008071 JAS61698.1 GBBI01004211 JAC14501.1 LNIX01000036 OXA39969.1 LNIX01000016 OXA46198.1 LNIX01000074 OXA36769.1 GDHC01006469 JAQ12160.1 ABLF02042806 GBHO01007689 JAG35915.1 ABLF02012239 ABLF02012241 ABLF02017372 ABLF02042178 ABLF02066436 ABLF02036131 ABLF02041527 ABLF02064049 AXCM01013330 ABLF02005251 ABLF02055296 GBHO01039838 GBHO01006034 JAG03766.1 JAG37570.1 GFTR01008866 JAW07560.1 LNIX01000044 OXA38660.1 GGMR01017803 MBY30422.1 GGMR01010750 MBY23369.1 OXA51382.1 QOIP01000009 RLU19024.1 GGMR01018506 MBY31125.1 LNIX01000002 OXA59522.1 GDHC01014057 JAQ04572.1 KQ458473 KPJ05885.1 LBMM01010417 KMQ87475.1 GBHO01044316 JAF99287.1 GDHC01016272 JAQ02357.1 GDHC01014268 JAQ04361.1 GBHO01044317 GBHO01044315 JAF99286.1 JAF99288.1 GBHO01028371 GBHO01028370 JAG15233.1 JAG15234.1 ABLF02035780 LNIX01000001 OXA61293.1 LNIX01000003 OXA58579.1 GALX01003264 JAB65202.1
Proteomes
Pfam
Interpro
IPR008042
Retrotrans_Pao
+ More
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR040676 DUF5641
IPR008737 Peptidase_asp_put
IPR005312 DUF1759
IPR000477 RT_dom
IPR021109 Peptidase_aspartic_dom_sf
IPR011012 Longin-like_dom_sf
IPR010908 Longin_dom
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR041373 RT_RNaseH
IPR001969 Aspartic_peptidase_AS
IPR041577 RT_RNaseH_2
IPR001995 Peptidase_A2_cat
IPR017452 GPCR_Rhodpsn_7TM
IPR000276 GPCR_Rhodpsn
IPR000331 Rap_GAP_dom
IPR035974 Rap/Ran-GAP_sf
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR040676 DUF5641
IPR008737 Peptidase_asp_put
IPR005312 DUF1759
IPR000477 RT_dom
IPR021109 Peptidase_aspartic_dom_sf
IPR011012 Longin-like_dom_sf
IPR010908 Longin_dom
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR041373 RT_RNaseH
IPR001969 Aspartic_peptidase_AS
IPR041577 RT_RNaseH_2
IPR001995 Peptidase_A2_cat
IPR017452 GPCR_Rhodpsn_7TM
IPR000276 GPCR_Rhodpsn
IPR000331 Rap_GAP_dom
IPR035974 Rap/Ran-GAP_sf
SUPFAM
Gene 3D
ProteinModelPortal
A0A1S4EIX5
A0A1S3DJJ0
A0A1S3DJ34
A0A1S3DJC0
A0A1S3DIZ2
A0A023F096
+ More
A0A1S3DMK8 A0A1Y1LIC8 A0A1S3DMT1 A0A3Q0JCL2 A0A1S3DH93 A0A1S3DHM0 A0A3Q0J7N1 A0A3Q0J7K9 A0A3Q0JJJ7 A0A3Q0JFQ3 A0A3Q0J0P0 A0A1B6LLT3 A0A1B6MT91 A0A1B6KIX6 A0A1Y1LM33 A0A1Y1LIG5 A0A3Q0JBZ9 A0A437AV42 V5GHM7 A0A1Y1KMX0 X1WRA3 A0A1B6KZV2 A0A1B6MUZ9 X1WY95 A0A0J7K7U0 V5GC07 A0A2S2NCY5 J9JVD6 J9KD60 A0A2S2PM34 A0A0J7KDN6 X1XU26 X1XTX7 A0A0A9YYJ5 A0A2S2QPS5 A0A226CWA5 A0A087T9T6 A0A0J7KCR4 A0A1S3DGE3 J9JUK4 A0A2S2PNC2 J9JLG6 W8C1U1 J9K351 A0A224XG39 A0A226E226 A0A1B6GGX5 A0A023F0J1 A0A226D417 A0A226DMG9 A0A3Q0JI79 A0A1S3DHZ9 A0A1S3DIE2 A0A226CV54 A0A1S3DRF1 A0A3Q0JDB5 A0A146M0N4 X1WNM7 A0A0A9Z275 X1WY64 X1WZY4 A0A182MIR3 A0A1S3DM17 J9M7R2 A0A1U8N859 A0A0A9W8L8 A0A224XGH3 A0A226D0T3 A0A3Q0JP94 A0A2S2PLU2 A0A2S2P1P0 A0A226E0X4 A0A3L8DF49 A0A2S2PNZ4 A0A226EPI5 A0A146LBU6 A0A0N0PAG9 A0A0J7N465 A0A0A9VYL4 A0A146L5E8 A0A146L7F7 A0A0A9VSF7 A0A0A9X8U7 J9LPA9 A0A226EUK6 A0A226EM86 V5GU69
A0A1S3DMK8 A0A1Y1LIC8 A0A1S3DMT1 A0A3Q0JCL2 A0A1S3DH93 A0A1S3DHM0 A0A3Q0J7N1 A0A3Q0J7K9 A0A3Q0JJJ7 A0A3Q0JFQ3 A0A3Q0J0P0 A0A1B6LLT3 A0A1B6MT91 A0A1B6KIX6 A0A1Y1LM33 A0A1Y1LIG5 A0A3Q0JBZ9 A0A437AV42 V5GHM7 A0A1Y1KMX0 X1WRA3 A0A1B6KZV2 A0A1B6MUZ9 X1WY95 A0A0J7K7U0 V5GC07 A0A2S2NCY5 J9JVD6 J9KD60 A0A2S2PM34 A0A0J7KDN6 X1XU26 X1XTX7 A0A0A9YYJ5 A0A2S2QPS5 A0A226CWA5 A0A087T9T6 A0A0J7KCR4 A0A1S3DGE3 J9JUK4 A0A2S2PNC2 J9JLG6 W8C1U1 J9K351 A0A224XG39 A0A226E226 A0A1B6GGX5 A0A023F0J1 A0A226D417 A0A226DMG9 A0A3Q0JI79 A0A1S3DHZ9 A0A1S3DIE2 A0A226CV54 A0A1S3DRF1 A0A3Q0JDB5 A0A146M0N4 X1WNM7 A0A0A9Z275 X1WY64 X1WZY4 A0A182MIR3 A0A1S3DM17 J9M7R2 A0A1U8N859 A0A0A9W8L8 A0A224XGH3 A0A226D0T3 A0A3Q0JP94 A0A2S2PLU2 A0A2S2P1P0 A0A226E0X4 A0A3L8DF49 A0A2S2PNZ4 A0A226EPI5 A0A146LBU6 A0A0N0PAG9 A0A0J7N465 A0A0A9VYL4 A0A146L5E8 A0A146L7F7 A0A0A9VSF7 A0A0A9X8U7 J9LPA9 A0A226EUK6 A0A226EM86 V5GU69
Ontologies
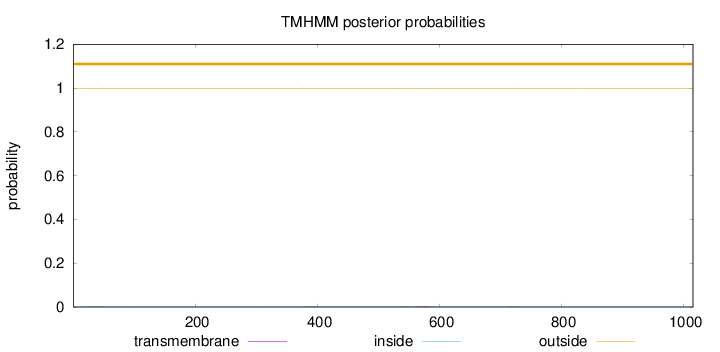
Topology
Length:
1015
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0891
Exp number, first 60 AAs:
0.01558
Total prob of N-in:
0.00121
outside
1 - 1015
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
158.052143
CSRT
0
Interpretation
Uncertain
