Gene
KWMTBOMO09681
Pre Gene Modal
BGIBMGA012957
Annotation
PREDICTED:_uncharacterized_protein_LOC106716472?_partial_[Papilio_machaon]
Location in the cell
Mitochondrial Reliability : 1.327
Sequence
CDS
ATGGCGTTGCGACTTAAAAAGGATATCAAGAAAGCGTCGTATTACGTTTGGTTCCTCGGGGCTCAGGAATCTCGCGGTCTGCGGGGCGAGGAGTTTGCGCTACCGGCGATCACGTTACTCGAGGAGAGAGCAAGGGAGTTGGAACCCTTCAAGGTTACGTTGCAGGTCTCTCTATGTTCTAAAGTTATCTCAGTCACAAGGCTTGTTGTATAA
Protein
MALRLKKDIKKASYYVWFLGAQESRGLRGEEFALPAITLLEERARELEPFKVTLQVSLCSKVISVTRLVV
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
S4NMH1
A0A1E1WIH4
A0A2A4J4A8
A0A2A4J328
H9JTU5
A0A194PZY2
+ More
A0A194R2T9 A0A212FBQ0 A0A3S2LHQ9 A0A2H1VCR9 A0A0L7KV33 D2A4Q8 A0A067RHZ4 A0A2J7PDV2 N6SXH9 A0A1Y1KCI1 U4UQ59 A0A2P8XZV1 A0A336KP94 A0A443SML7 A0A2M4CXP0 A0A1W4XIC0 A0A336K9P7 A0A1S4EJH7 A0A1B0CEV2 A0A1B6E327 A0A182NZ14 A0A3Q0JBV0 A0A182II21 A0A1I8JVN9 A0A453YN19 B0W6Q9 A0ND11 A0A1Q3FYB1 A0A182G446 A0A1W7R4V0 A0A023EHT2 A0A1B0DLC4 A0A182Y3Z6 A0A340TB91 A0A087TJX2 E0VH39 T1DSM8 A0A182FQY5 A0A182PCK8 A0A182TI08 A0A182R446 A0A182VZT5 A0A182SYL2 A0A1A9WSX6 A0A182MC88 A0A1A9VRS3 A0A1B0FDX3 A0A1A9XH55 A0A1B0AN89 A0A1A9ZIB9 T1PNZ1 A0A182Q4K2 A0A182IRG7 A0A2L2YZR6 B4NPZ2 A0A1I8PD26 B3NUK4 B3MZH5 A0A1I8M2N1 A0A1W4UQP5 B4Q006 B4GVJ1 B4R6S0 B4IJT4 A0A0L0C2A6 A0A0K8W462 B5DN89 A0A3B0K2V7 B4JNU0 A0A0K8V8R7 A0A0K8WJB3 B4L3K3 A0A0K8V3C8 Q9W3B8 A0A0A1X4R6 A0A0A1WLA1 A0A034V5M0 A0A132A0Y9 M9PE29 A0A0M5J1D9 A0A1J1HEI2 B4MD91 W8ATN1 T1J2M3 B7QGS3 J9KNI7 A0A2S2QAT3 A0A2S2P1C6 A0A1Y3BB07 A0A154P7T6 T1JUH2 A0A088AMP8 A0A2A3E448 A0A0L7R385
A0A194R2T9 A0A212FBQ0 A0A3S2LHQ9 A0A2H1VCR9 A0A0L7KV33 D2A4Q8 A0A067RHZ4 A0A2J7PDV2 N6SXH9 A0A1Y1KCI1 U4UQ59 A0A2P8XZV1 A0A336KP94 A0A443SML7 A0A2M4CXP0 A0A1W4XIC0 A0A336K9P7 A0A1S4EJH7 A0A1B0CEV2 A0A1B6E327 A0A182NZ14 A0A3Q0JBV0 A0A182II21 A0A1I8JVN9 A0A453YN19 B0W6Q9 A0ND11 A0A1Q3FYB1 A0A182G446 A0A1W7R4V0 A0A023EHT2 A0A1B0DLC4 A0A182Y3Z6 A0A340TB91 A0A087TJX2 E0VH39 T1DSM8 A0A182FQY5 A0A182PCK8 A0A182TI08 A0A182R446 A0A182VZT5 A0A182SYL2 A0A1A9WSX6 A0A182MC88 A0A1A9VRS3 A0A1B0FDX3 A0A1A9XH55 A0A1B0AN89 A0A1A9ZIB9 T1PNZ1 A0A182Q4K2 A0A182IRG7 A0A2L2YZR6 B4NPZ2 A0A1I8PD26 B3NUK4 B3MZH5 A0A1I8M2N1 A0A1W4UQP5 B4Q006 B4GVJ1 B4R6S0 B4IJT4 A0A0L0C2A6 A0A0K8W462 B5DN89 A0A3B0K2V7 B4JNU0 A0A0K8V8R7 A0A0K8WJB3 B4L3K3 A0A0K8V3C8 Q9W3B8 A0A0A1X4R6 A0A0A1WLA1 A0A034V5M0 A0A132A0Y9 M9PE29 A0A0M5J1D9 A0A1J1HEI2 B4MD91 W8ATN1 T1J2M3 B7QGS3 J9KNI7 A0A2S2QAT3 A0A2S2P1C6 A0A1Y3BB07 A0A154P7T6 T1JUH2 A0A088AMP8 A0A2A3E448 A0A0L7R385
Pubmed
23622113
19121390
26354079
22118469
26227816
18362917
+ More
19820115 24845553 23537049 28004739 29403074 20966253 12364791 14747013 17210077 26483478 24945155 25244985 20566863 26561354 17994087 18057021 25315136 17550304 26108605 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 25348373 26555130 24495485
19820115 24845553 23537049 28004739 29403074 20966253 12364791 14747013 17210077 26483478 24945155 25244985 20566863 26561354 17994087 18057021 25315136 17550304 26108605 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 25348373 26555130 24495485
EMBL
GAIX01012669
JAA79891.1
GDQN01004357
JAT86697.1
NWSH01003446
PCG66280.1
+ More
PCG66279.1 BABH01003631 KQ459582 KPI98887.1 KQ460878 KPJ11560.1 AGBW02009293 OWR51165.1 RSAL01000115 RVE46891.1 ODYU01001659 SOQ38192.1 JTDY01005501 KOB66931.1 KQ971344 EFA05191.2 KK852622 KDR20004.1 NEVH01026140 PNF14502.1 APGK01052848 KB741216 ENN72474.1 GEZM01086684 JAV59064.1 KI210229 ERL96204.1 PYGN01001105 PSN37520.1 UFQS01000659 UFQT01000659 SSX05862.1 SSX26221.1 NCKV01001230 RWS28758.1 GGFL01005909 MBW70087.1 UFQS01000150 UFQT01000150 SSX00522.1 SSX20902.1 AJWK01009289 AJWK01009290 AJWK01009291 AJWK01009292 AJWK01009293 AJWK01009294 AJWK01009295 AJWK01009296 AJWK01009297 AJWK01009298 GEDC01013282 GEDC01004977 JAS24016.1 JAS32321.1 APCN01000245 DS231849 EDS36890.1 AAAB01008859 EAU77095.3 GFDL01002522 JAV32523.1 JXUM01041988 JXUM01041989 KQ561306 KXJ79050.1 GEHC01001471 JAV46174.1 GAPW01004981 JAC08617.1 AJVK01016377 AJVK01016378 KK115557 KFM65411.1 DS235158 EEB12695.1 GAMD01000130 JAB01461.1 AXCM01000144 CCAG010011618 JXJN01000753 JXJN01000754 KA650466 AFP65095.1 AXCN02001476 IAAA01085888 LAA13566.1 CH964291 EDW86217.2 CH954180 EDV46327.1 CH902635 EDV33776.1 KPU74744.1 CM000162 EDX02212.1 CH479192 EDW26478.1 CM000366 EDX17450.1 CH480849 EDW51237.1 JRES01000984 KNC26455.1 GDHF01027233 GDHF01015234 GDHF01009814 GDHF01007968 GDHF01006470 JAI25081.1 JAI37080.1 JAI42500.1 JAI44346.1 JAI45844.1 CH379064 EDY72896.1 OUUW01000015 SPP88615.1 CH916371 EDV92383.1 GDHF01017027 JAI35287.1 GDHF01001096 JAI51218.1 CH933810 EDW07131.2 GDHF01018963 JAI33351.1 AE014298 BT032850 AAF46414.3 ACD81864.1 GBXI01008170 GBXI01000240 JAD06122.1 JAD14052.1 GBXI01015565 GBXI01015104 GBXI01001580 JAC98726.1 JAC99187.1 JAD12712.1 GAKP01021518 GAKP01021517 GAKP01021516 GAKP01021515 JAC37437.1 JXLN01009611 KPM04627.1 AGB95202.1 CP012528 ALC49644.1 CVRI01000001 CRK86243.1 CH940660 EDW58163.1 KRF78154.1 KRF78155.1 GAMC01018557 JAB87998.1 JH431806 ABJB010552123 DS933998 EEC18045.1 ABLF02024892 ABLF02024893 ABLF02024895 ABLF02024896 ABLF02024897 ABLF02037090 ABLF02066510 GGMS01005645 MBY74848.1 GGMR01010409 MBY23028.1 MUJZ01037701 OTF76395.1 KQ434839 KZC07986.1 CAEY01000779 KZ288382 PBC26507.1 KQ414663 KOC65241.1
PCG66279.1 BABH01003631 KQ459582 KPI98887.1 KQ460878 KPJ11560.1 AGBW02009293 OWR51165.1 RSAL01000115 RVE46891.1 ODYU01001659 SOQ38192.1 JTDY01005501 KOB66931.1 KQ971344 EFA05191.2 KK852622 KDR20004.1 NEVH01026140 PNF14502.1 APGK01052848 KB741216 ENN72474.1 GEZM01086684 JAV59064.1 KI210229 ERL96204.1 PYGN01001105 PSN37520.1 UFQS01000659 UFQT01000659 SSX05862.1 SSX26221.1 NCKV01001230 RWS28758.1 GGFL01005909 MBW70087.1 UFQS01000150 UFQT01000150 SSX00522.1 SSX20902.1 AJWK01009289 AJWK01009290 AJWK01009291 AJWK01009292 AJWK01009293 AJWK01009294 AJWK01009295 AJWK01009296 AJWK01009297 AJWK01009298 GEDC01013282 GEDC01004977 JAS24016.1 JAS32321.1 APCN01000245 DS231849 EDS36890.1 AAAB01008859 EAU77095.3 GFDL01002522 JAV32523.1 JXUM01041988 JXUM01041989 KQ561306 KXJ79050.1 GEHC01001471 JAV46174.1 GAPW01004981 JAC08617.1 AJVK01016377 AJVK01016378 KK115557 KFM65411.1 DS235158 EEB12695.1 GAMD01000130 JAB01461.1 AXCM01000144 CCAG010011618 JXJN01000753 JXJN01000754 KA650466 AFP65095.1 AXCN02001476 IAAA01085888 LAA13566.1 CH964291 EDW86217.2 CH954180 EDV46327.1 CH902635 EDV33776.1 KPU74744.1 CM000162 EDX02212.1 CH479192 EDW26478.1 CM000366 EDX17450.1 CH480849 EDW51237.1 JRES01000984 KNC26455.1 GDHF01027233 GDHF01015234 GDHF01009814 GDHF01007968 GDHF01006470 JAI25081.1 JAI37080.1 JAI42500.1 JAI44346.1 JAI45844.1 CH379064 EDY72896.1 OUUW01000015 SPP88615.1 CH916371 EDV92383.1 GDHF01017027 JAI35287.1 GDHF01001096 JAI51218.1 CH933810 EDW07131.2 GDHF01018963 JAI33351.1 AE014298 BT032850 AAF46414.3 ACD81864.1 GBXI01008170 GBXI01000240 JAD06122.1 JAD14052.1 GBXI01015565 GBXI01015104 GBXI01001580 JAC98726.1 JAC99187.1 JAD12712.1 GAKP01021518 GAKP01021517 GAKP01021516 GAKP01021515 JAC37437.1 JXLN01009611 KPM04627.1 AGB95202.1 CP012528 ALC49644.1 CVRI01000001 CRK86243.1 CH940660 EDW58163.1 KRF78154.1 KRF78155.1 GAMC01018557 JAB87998.1 JH431806 ABJB010552123 DS933998 EEC18045.1 ABLF02024892 ABLF02024893 ABLF02024895 ABLF02024896 ABLF02024897 ABLF02037090 ABLF02066510 GGMS01005645 MBY74848.1 GGMR01010409 MBY23028.1 MUJZ01037701 OTF76395.1 KQ434839 KZC07986.1 CAEY01000779 KZ288382 PBC26507.1 KQ414663 KOC65241.1
Proteomes
UP000218220
UP000005204
UP000053268
UP000053240
UP000007151
UP000283053
+ More
UP000037510 UP000007266 UP000027135 UP000235965 UP000019118 UP000030742 UP000245037 UP000288716 UP000192223 UP000079169 UP000092461 UP000075884 UP000075840 UP000076407 UP000075882 UP000002320 UP000007062 UP000069940 UP000249989 UP000092462 UP000076408 UP000075903 UP000054359 UP000009046 UP000069272 UP000075885 UP000075902 UP000075900 UP000075920 UP000075901 UP000091820 UP000075883 UP000078200 UP000092444 UP000092443 UP000092460 UP000092445 UP000075886 UP000075880 UP000007798 UP000095300 UP000008711 UP000007801 UP000095301 UP000192221 UP000002282 UP000008744 UP000000304 UP000001292 UP000037069 UP000001819 UP000268350 UP000001070 UP000009192 UP000000803 UP000092553 UP000183832 UP000008792 UP000001555 UP000007819 UP000076502 UP000015104 UP000005203 UP000242457 UP000053825
UP000037510 UP000007266 UP000027135 UP000235965 UP000019118 UP000030742 UP000245037 UP000288716 UP000192223 UP000079169 UP000092461 UP000075884 UP000075840 UP000076407 UP000075882 UP000002320 UP000007062 UP000069940 UP000249989 UP000092462 UP000076408 UP000075903 UP000054359 UP000009046 UP000069272 UP000075885 UP000075902 UP000075900 UP000075920 UP000075901 UP000091820 UP000075883 UP000078200 UP000092444 UP000092443 UP000092460 UP000092445 UP000075886 UP000075880 UP000007798 UP000095300 UP000008711 UP000007801 UP000095301 UP000192221 UP000002282 UP000008744 UP000000304 UP000001292 UP000037069 UP000001819 UP000268350 UP000001070 UP000009192 UP000000803 UP000092553 UP000183832 UP000008792 UP000001555 UP000007819 UP000076502 UP000015104 UP000005203 UP000242457 UP000053825
Interpro
Gene 3D
CDD
ProteinModelPortal
S4NMH1
A0A1E1WIH4
A0A2A4J4A8
A0A2A4J328
H9JTU5
A0A194PZY2
+ More
A0A194R2T9 A0A212FBQ0 A0A3S2LHQ9 A0A2H1VCR9 A0A0L7KV33 D2A4Q8 A0A067RHZ4 A0A2J7PDV2 N6SXH9 A0A1Y1KCI1 U4UQ59 A0A2P8XZV1 A0A336KP94 A0A443SML7 A0A2M4CXP0 A0A1W4XIC0 A0A336K9P7 A0A1S4EJH7 A0A1B0CEV2 A0A1B6E327 A0A182NZ14 A0A3Q0JBV0 A0A182II21 A0A1I8JVN9 A0A453YN19 B0W6Q9 A0ND11 A0A1Q3FYB1 A0A182G446 A0A1W7R4V0 A0A023EHT2 A0A1B0DLC4 A0A182Y3Z6 A0A340TB91 A0A087TJX2 E0VH39 T1DSM8 A0A182FQY5 A0A182PCK8 A0A182TI08 A0A182R446 A0A182VZT5 A0A182SYL2 A0A1A9WSX6 A0A182MC88 A0A1A9VRS3 A0A1B0FDX3 A0A1A9XH55 A0A1B0AN89 A0A1A9ZIB9 T1PNZ1 A0A182Q4K2 A0A182IRG7 A0A2L2YZR6 B4NPZ2 A0A1I8PD26 B3NUK4 B3MZH5 A0A1I8M2N1 A0A1W4UQP5 B4Q006 B4GVJ1 B4R6S0 B4IJT4 A0A0L0C2A6 A0A0K8W462 B5DN89 A0A3B0K2V7 B4JNU0 A0A0K8V8R7 A0A0K8WJB3 B4L3K3 A0A0K8V3C8 Q9W3B8 A0A0A1X4R6 A0A0A1WLA1 A0A034V5M0 A0A132A0Y9 M9PE29 A0A0M5J1D9 A0A1J1HEI2 B4MD91 W8ATN1 T1J2M3 B7QGS3 J9KNI7 A0A2S2QAT3 A0A2S2P1C6 A0A1Y3BB07 A0A154P7T6 T1JUH2 A0A088AMP8 A0A2A3E448 A0A0L7R385
A0A194R2T9 A0A212FBQ0 A0A3S2LHQ9 A0A2H1VCR9 A0A0L7KV33 D2A4Q8 A0A067RHZ4 A0A2J7PDV2 N6SXH9 A0A1Y1KCI1 U4UQ59 A0A2P8XZV1 A0A336KP94 A0A443SML7 A0A2M4CXP0 A0A1W4XIC0 A0A336K9P7 A0A1S4EJH7 A0A1B0CEV2 A0A1B6E327 A0A182NZ14 A0A3Q0JBV0 A0A182II21 A0A1I8JVN9 A0A453YN19 B0W6Q9 A0ND11 A0A1Q3FYB1 A0A182G446 A0A1W7R4V0 A0A023EHT2 A0A1B0DLC4 A0A182Y3Z6 A0A340TB91 A0A087TJX2 E0VH39 T1DSM8 A0A182FQY5 A0A182PCK8 A0A182TI08 A0A182R446 A0A182VZT5 A0A182SYL2 A0A1A9WSX6 A0A182MC88 A0A1A9VRS3 A0A1B0FDX3 A0A1A9XH55 A0A1B0AN89 A0A1A9ZIB9 T1PNZ1 A0A182Q4K2 A0A182IRG7 A0A2L2YZR6 B4NPZ2 A0A1I8PD26 B3NUK4 B3MZH5 A0A1I8M2N1 A0A1W4UQP5 B4Q006 B4GVJ1 B4R6S0 B4IJT4 A0A0L0C2A6 A0A0K8W462 B5DN89 A0A3B0K2V7 B4JNU0 A0A0K8V8R7 A0A0K8WJB3 B4L3K3 A0A0K8V3C8 Q9W3B8 A0A0A1X4R6 A0A0A1WLA1 A0A034V5M0 A0A132A0Y9 M9PE29 A0A0M5J1D9 A0A1J1HEI2 B4MD91 W8ATN1 T1J2M3 B7QGS3 J9KNI7 A0A2S2QAT3 A0A2S2P1C6 A0A1Y3BB07 A0A154P7T6 T1JUH2 A0A088AMP8 A0A2A3E448 A0A0L7R385
Ontologies
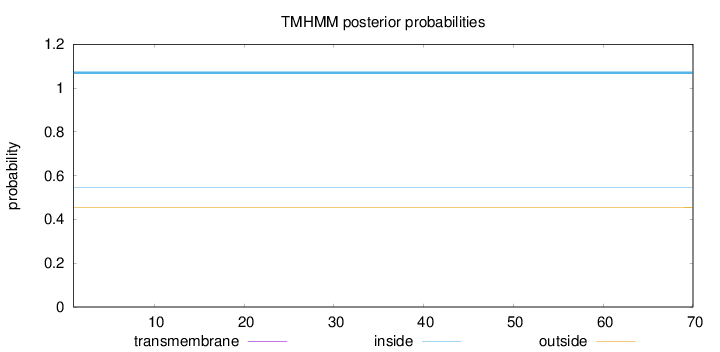
Topology
Length:
70
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00432
Exp number, first 60 AAs:
0.0031
Total prob of N-in:
0.54604
inside
1 - 70
Population Genetic Test Statistics
Pi
221.173988
Theta
160.71661
Tajima's D
1.099431
CLR
0.000046
CSRT
0.685915704214789
Interpretation
Uncertain
