Gene
KWMTBOMO09676
Annotation
red_protein_[Danaus_plexippus]
Location in the cell
Nuclear Reliability : 4.439
Sequence
CDS
ATGGAAGATCACAGGGGTGTGGAGGACACACCGGCTTCACAGAGGCTTACAAATGATGACTTTAGGAAATTACTGATGACTCCAAGATCTACTCCTTCAGGAGGGACACACGCTCCTGGCTCCATTCGAGAGGCTATGGCGAATGCAGGATCCATGCCACCACCATCAGAGAATAAAGGAGAGTTACGTCGTAAGAAAAAGTCATACTATGCTGCTCTTAAGAAACAAGAAGACAACAAGCTAGCAGAGCTTGCGGAGAAATACAGAGACAGAGCTAGAGAGAGGAGAGATGGTGTGAATGATTTAAGTGTTATTGATCCAACAAGCAACACAAGTAGTGCCTATAGAGCTGTTGCTCCAGATTTGAAATCAGGTCTAGATGCCAAAGAAAGGAGGAGACAACTTATCCAGGAATCCAAATATCTTGGTGGTGACATGGAGCACACTCACTTGGTAAAAGGTCTCGATTATGCTTTGTTGCAGAAGGTTCGTTCAGAAATACAAACTCGGGAGCAAGAAGAAGAAGCCGAAATGGAGAGACTGGTCACTATTCCCGTGGAACCTGTAAAAGAAAAGAAAGAAGAAAAGAAGGAAGTTGAAGAAGAGATGCAGTTCAAGACTCAAATGGGAAAAAATATTTTTGACATGATTCTAGAACAAAAGAACAAGAAAGTTGTCCGCAGCGAAATGTTTGCTCCCGGTCGTATGGCGTACGTGGTCGAGCTCGACGAAGAAGGCACAATCGATTCCGATATACCCACAACGTTGACTCGGAGTAAAGCGGACGTTCCCGAAATGGACGAAAGGAGTTCGGCTTCGAGTGCAAATGATCTTGTCGTCGAGAAACTGACTCAGATCTTCTCATATTTGAGGCACGGAAGACATAAGAAGCTGAAGAAGACTAAGGACAAGACGACGGAGAAGGGTCGTAACGACGACTCTATATACGGCGAGATTGGTGACTACATGCCGGGAGAACGCCGCTCCGACAGGAGGGAGGAGAGGCCGCGAACCGGATACTTTGATAAGCCTGTTGAAACCGAGAAGGAAGAGGGCCCGATCCCTCGTCGTCAAGCAACAGGCAATAACGAAGAGCGCGACAAACGTTCCGCGGCGCTGCTGTCCAAGCTGGCGGCTGAGCCCGAGGGCTACGCCGAGTGCTACCCCGGCCTCAGGGAGATGGACGACGCCATAGACGACTCCGACGATGAAGTCGACTACACCAAAATGGACGCCGGGAACAAGAAGGGACCGATCGGTCGCTGGGATTTCGACACGCAAGAGGAATACTCCGCCTACATGAGCAGCAAAGAGGCGTTACCTAAGGCGGCCTTCCAATATGGCGTCAAAACGCAAGACGGACGGAAAACAAGGAAAACGAAGGACAAAAGCGAAAAGGCAGAGCTGGACAGAGAATGGCAACAGATTCAAAACATCATACAGAAACGAAAAGCGCCTCTGCATTCTGACGAACCAGGAGTTAAAGCCTCAAAATATTAA
Protein
MEDHRGVEDTPASQRLTNDDFRKLLMTPRSTPSGGTHAPGSIREAMANAGSMPPPSENKGELRRKKKSYYAALKKQEDNKLAELAEKYRDRARERRDGVNDLSVIDPTSNTSSAYRAVAPDLKSGLDAKERRRQLIQESKYLGGDMEHTHLVKGLDYALLQKVRSEIQTREQEEEAEMERLVTIPVEPVKEKKEEKKEVEEEMQFKTQMGKNIFDMILEQKNKKVVRSEMFAPGRMAYVVELDEEGTIDSDIPTTLTRSKADVPEMDERSSASSANDLVVEKLTQIFSYLRHGRHKKLKKTKDKTTEKGRNDDSIYGEIGDYMPGERRSDRREERPRTGYFDKPVETEKEEGPIPRRQATGNNEERDKRSAALLSKLAAEPEGYAECYPGLREMDDAIDDSDDEVDYTKMDAGNKKGPIGRWDFDTQEEYSAYMSSKEALPKAAFQYGVKTQDGRKTRKTKDKSEKAELDREWQQIQNIIQKRKAPLHSDEPGVKASKY
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
A0A3S2TIG3
A0A194PZY7
S4PPI7
A0A212FBP2
A0A2A4K8P4
A0A1Y1JUZ9
+ More
A0A194R6R5 D2A510 A0A0T6AU23 A0A1W4WYZ7 A0A182G740 A0A182JVE7 B0W646 Q16V55 A0A1S4FN34 U4UQY5 A0A084VZT7 A0A182JCS5 A0A182TTQ9 Q7PMU2 A0A182LI88 A0A0L7R0R3 N6TSY6 A0A2M4AS08 A0A182TC88 A0A087ZYB6 K7J6P0 A0A2A3EED5 A0A232EDW6 A0A0J7KVB9 A0A1Q3F8N9 A0A154PHX3 A0A151IHH8 A0A0N0BH30 E2BWP8 F4W7E2 A0A151JTC6 A0A182YI49 A0A158NPV2 E9IC81 A0A195DTR1 A0A151X8N3 A0A195ATG3 A0A026WTZ5 A0A182MIN6 E2A3S3 W5JQU4 E0VKU6 A0A182NNA6 A0A182QEM8 A0A182P610 A0A182FLU3 A0A182R2R9 A0A182WAR8 A0A2R7VWY3 A0A2P8XP32 A0A1B6FD66 A0A0A9Z548 A0A069DTY5 U5EYM4 A0A1B6H9K6 J9JV79 A0A1L8DLT4 A0A2S2NYZ1 A0A023F432 A0A0V0GBQ9 A0A224XMP1 A0A1I8Q213 A0A2J7QAM8 A0A2R7VRD3 A0A2H8TDV2 A0A2M4BKU8 A0A1B6D3C8 A0A336LEX5 A0A0P4VS43 A0A2M4BKV1 T1HPM5 A0A067QJX1 A0A336N0T3 A0A1I8M387 A0A0L0BN98 A0A034VVS6 A0A0K8WFM1 W8BTG0 A0A1A9V7X1 A0A1B0FMC9 A0A1A9Z6I3 B4N996 B3P1T7 A0A0L7L113 A0A1B0FDM0 A0A1S4EJS9 B4HKI0 B4QXA1 A0A1A9V0D2 A0A023GLA1 G3MNK3 B3M1Z4 L7MAC3 A0A1E1XCE3
A0A194R6R5 D2A510 A0A0T6AU23 A0A1W4WYZ7 A0A182G740 A0A182JVE7 B0W646 Q16V55 A0A1S4FN34 U4UQY5 A0A084VZT7 A0A182JCS5 A0A182TTQ9 Q7PMU2 A0A182LI88 A0A0L7R0R3 N6TSY6 A0A2M4AS08 A0A182TC88 A0A087ZYB6 K7J6P0 A0A2A3EED5 A0A232EDW6 A0A0J7KVB9 A0A1Q3F8N9 A0A154PHX3 A0A151IHH8 A0A0N0BH30 E2BWP8 F4W7E2 A0A151JTC6 A0A182YI49 A0A158NPV2 E9IC81 A0A195DTR1 A0A151X8N3 A0A195ATG3 A0A026WTZ5 A0A182MIN6 E2A3S3 W5JQU4 E0VKU6 A0A182NNA6 A0A182QEM8 A0A182P610 A0A182FLU3 A0A182R2R9 A0A182WAR8 A0A2R7VWY3 A0A2P8XP32 A0A1B6FD66 A0A0A9Z548 A0A069DTY5 U5EYM4 A0A1B6H9K6 J9JV79 A0A1L8DLT4 A0A2S2NYZ1 A0A023F432 A0A0V0GBQ9 A0A224XMP1 A0A1I8Q213 A0A2J7QAM8 A0A2R7VRD3 A0A2H8TDV2 A0A2M4BKU8 A0A1B6D3C8 A0A336LEX5 A0A0P4VS43 A0A2M4BKV1 T1HPM5 A0A067QJX1 A0A336N0T3 A0A1I8M387 A0A0L0BN98 A0A034VVS6 A0A0K8WFM1 W8BTG0 A0A1A9V7X1 A0A1B0FMC9 A0A1A9Z6I3 B4N996 B3P1T7 A0A0L7L113 A0A1B0FDM0 A0A1S4EJS9 B4HKI0 B4QXA1 A0A1A9V0D2 A0A023GLA1 G3MNK3 B3M1Z4 L7MAC3 A0A1E1XCE3
Pubmed
26354079
23622113
22118469
28004739
18362917
19820115
+ More
26483478 17510324 23537049 24438588 12364791 14747013 17210077 20966253 20075255 28648823 20798317 21719571 25244985 21347285 21282665 24508170 30249741 20920257 23761445 20566863 29403074 25401762 26823975 26334808 25474469 27129103 24845553 25315136 26108605 25348373 24495485 17994087 26227816 22216098 25576852 28503490
26483478 17510324 23537049 24438588 12364791 14747013 17210077 20966253 20075255 28648823 20798317 21719571 25244985 21347285 21282665 24508170 30249741 20920257 23761445 20566863 29403074 25401762 26823975 26334808 25474469 27129103 24845553 25315136 26108605 25348373 24495485 17994087 26227816 22216098 25576852 28503490
EMBL
RSAL01000115
RVE46888.1
KQ459582
KPI98892.1
GAIX01000222
JAA92338.1
+ More
AGBW02009293 OWR51161.1 NWSH01000064 PCG80032.1 GEZM01102094 JAV51978.1 KQ460878 KPJ11556.1 KQ971361 EFA05295.1 LJIG01022804 KRT78625.1 JXUM01046077 JXUM01046078 JXUM01046079 JXUM01046080 JXUM01046081 JXUM01046082 JXUM01046083 JXUM01046084 JXUM01046085 JXUM01046086 KQ561462 KXJ78499.1 DS231847 EDS36393.1 CH477603 EAT38415.1 KB632330 ERL92566.1 ATLV01018958 KE525256 KFB43481.1 AAAB01008968 EAA13204.5 KQ414670 KOC64381.1 APGK01055721 KB741269 ENN71461.1 GGFK01010201 MBW43522.1 KZ288266 PBC30115.1 NNAY01005839 OXU16533.1 LBMM01002883 KMQ94248.1 GFDL01011111 JAV23934.1 KQ434912 KZC11397.1 KQ977606 KYN01417.1 KQ435760 KOX75638.1 GL451165 EFN79871.1 GL887844 EGI69815.1 KQ982014 KYN30627.1 ADTU01022757 GL762206 EFZ21826.1 KQ980487 KYN15914.1 KQ982409 KYQ56731.1 KQ976745 KYM75481.1 KK107105 QOIP01000011 EZA59492.1 RLU16407.1 AXCM01000074 GL436457 EFN71965.1 ADMH02000702 ETN65280.1 DS235250 EEB14002.1 AXCN02001904 KK854140 PTY11972.1 PYGN01001609 PSN33760.1 GECZ01021634 JAS48135.1 GBHO01005146 GBRD01013489 GDHC01003392 JAG38458.1 JAG52337.1 JAQ15237.1 GBGD01001321 JAC87568.1 GANO01000259 JAB59612.1 GECU01036298 JAS71408.1 ABLF02026148 GFDF01006665 JAV07419.1 GGMR01009766 MBY22385.1 GBBI01002496 JAC16216.1 GECL01000847 JAP05277.1 GFTR01006574 JAW09852.1 NEVH01016329 PNF25632.1 KK854026 PTY09698.1 GFXV01000481 MBW12286.1 GGFJ01004555 MBW53696.1 GEDC01017118 JAS20180.1 UFQS01004251 UFQT01004251 SSX16280.1 SSX35600.1 GDKW01001336 JAI55259.1 GGFJ01004556 MBW53697.1 ACPB03017125 KK853383 KDR07997.1 UFQS01003096 UFQT01003096 SSX15177.1 SSX34553.1 JRES01001619 KNC21497.1 GAKP01013337 GAKP01013332 GAKP01013329 JAC45623.1 GDHF01024848 GDHF01021453 GDHF01002659 JAI27466.1 JAI30861.1 JAI49655.1 GAMC01010042 GAMC01010039 GAMC01010037 JAB96516.1 CCAG010018533 CH964232 EDW80529.1 CH954181 EDV49825.1 JTDY01003784 KOB69011.1 CCAG010009795 CH480815 EDW42930.1 CM000364 EDX13672.1 GBBM01000814 JAC34604.1 JO843454 AEO35071.1 CH902617 EDV43318.1 GACK01004946 JAA60088.1 GFAC01002310 JAT96878.1
AGBW02009293 OWR51161.1 NWSH01000064 PCG80032.1 GEZM01102094 JAV51978.1 KQ460878 KPJ11556.1 KQ971361 EFA05295.1 LJIG01022804 KRT78625.1 JXUM01046077 JXUM01046078 JXUM01046079 JXUM01046080 JXUM01046081 JXUM01046082 JXUM01046083 JXUM01046084 JXUM01046085 JXUM01046086 KQ561462 KXJ78499.1 DS231847 EDS36393.1 CH477603 EAT38415.1 KB632330 ERL92566.1 ATLV01018958 KE525256 KFB43481.1 AAAB01008968 EAA13204.5 KQ414670 KOC64381.1 APGK01055721 KB741269 ENN71461.1 GGFK01010201 MBW43522.1 KZ288266 PBC30115.1 NNAY01005839 OXU16533.1 LBMM01002883 KMQ94248.1 GFDL01011111 JAV23934.1 KQ434912 KZC11397.1 KQ977606 KYN01417.1 KQ435760 KOX75638.1 GL451165 EFN79871.1 GL887844 EGI69815.1 KQ982014 KYN30627.1 ADTU01022757 GL762206 EFZ21826.1 KQ980487 KYN15914.1 KQ982409 KYQ56731.1 KQ976745 KYM75481.1 KK107105 QOIP01000011 EZA59492.1 RLU16407.1 AXCM01000074 GL436457 EFN71965.1 ADMH02000702 ETN65280.1 DS235250 EEB14002.1 AXCN02001904 KK854140 PTY11972.1 PYGN01001609 PSN33760.1 GECZ01021634 JAS48135.1 GBHO01005146 GBRD01013489 GDHC01003392 JAG38458.1 JAG52337.1 JAQ15237.1 GBGD01001321 JAC87568.1 GANO01000259 JAB59612.1 GECU01036298 JAS71408.1 ABLF02026148 GFDF01006665 JAV07419.1 GGMR01009766 MBY22385.1 GBBI01002496 JAC16216.1 GECL01000847 JAP05277.1 GFTR01006574 JAW09852.1 NEVH01016329 PNF25632.1 KK854026 PTY09698.1 GFXV01000481 MBW12286.1 GGFJ01004555 MBW53696.1 GEDC01017118 JAS20180.1 UFQS01004251 UFQT01004251 SSX16280.1 SSX35600.1 GDKW01001336 JAI55259.1 GGFJ01004556 MBW53697.1 ACPB03017125 KK853383 KDR07997.1 UFQS01003096 UFQT01003096 SSX15177.1 SSX34553.1 JRES01001619 KNC21497.1 GAKP01013337 GAKP01013332 GAKP01013329 JAC45623.1 GDHF01024848 GDHF01021453 GDHF01002659 JAI27466.1 JAI30861.1 JAI49655.1 GAMC01010042 GAMC01010039 GAMC01010037 JAB96516.1 CCAG010018533 CH964232 EDW80529.1 CH954181 EDV49825.1 JTDY01003784 KOB69011.1 CCAG010009795 CH480815 EDW42930.1 CM000364 EDX13672.1 GBBM01000814 JAC34604.1 JO843454 AEO35071.1 CH902617 EDV43318.1 GACK01004946 JAA60088.1 GFAC01002310 JAT96878.1
Proteomes
UP000283053
UP000053268
UP000007151
UP000218220
UP000053240
UP000007266
+ More
UP000192223 UP000069940 UP000249989 UP000075881 UP000002320 UP000008820 UP000030742 UP000030765 UP000075880 UP000075902 UP000007062 UP000075882 UP000053825 UP000019118 UP000075901 UP000005203 UP000002358 UP000242457 UP000215335 UP000036403 UP000076502 UP000078542 UP000053105 UP000008237 UP000007755 UP000078541 UP000076408 UP000005205 UP000078492 UP000075809 UP000078540 UP000053097 UP000279307 UP000075883 UP000000311 UP000000673 UP000009046 UP000075884 UP000075886 UP000075885 UP000069272 UP000075900 UP000075920 UP000245037 UP000007819 UP000095300 UP000235965 UP000015103 UP000027135 UP000095301 UP000037069 UP000078200 UP000092444 UP000092445 UP000007798 UP000008711 UP000037510 UP000079169 UP000001292 UP000000304 UP000007801
UP000192223 UP000069940 UP000249989 UP000075881 UP000002320 UP000008820 UP000030742 UP000030765 UP000075880 UP000075902 UP000007062 UP000075882 UP000053825 UP000019118 UP000075901 UP000005203 UP000002358 UP000242457 UP000215335 UP000036403 UP000076502 UP000078542 UP000053105 UP000008237 UP000007755 UP000078541 UP000076408 UP000005205 UP000078492 UP000075809 UP000078540 UP000053097 UP000279307 UP000075883 UP000000311 UP000000673 UP000009046 UP000075884 UP000075886 UP000075885 UP000069272 UP000075900 UP000075920 UP000245037 UP000007819 UP000095300 UP000235965 UP000015103 UP000027135 UP000095301 UP000037069 UP000078200 UP000092444 UP000092445 UP000007798 UP000008711 UP000037510 UP000079169 UP000001292 UP000000304 UP000007801
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A3S2TIG3
A0A194PZY7
S4PPI7
A0A212FBP2
A0A2A4K8P4
A0A1Y1JUZ9
+ More
A0A194R6R5 D2A510 A0A0T6AU23 A0A1W4WYZ7 A0A182G740 A0A182JVE7 B0W646 Q16V55 A0A1S4FN34 U4UQY5 A0A084VZT7 A0A182JCS5 A0A182TTQ9 Q7PMU2 A0A182LI88 A0A0L7R0R3 N6TSY6 A0A2M4AS08 A0A182TC88 A0A087ZYB6 K7J6P0 A0A2A3EED5 A0A232EDW6 A0A0J7KVB9 A0A1Q3F8N9 A0A154PHX3 A0A151IHH8 A0A0N0BH30 E2BWP8 F4W7E2 A0A151JTC6 A0A182YI49 A0A158NPV2 E9IC81 A0A195DTR1 A0A151X8N3 A0A195ATG3 A0A026WTZ5 A0A182MIN6 E2A3S3 W5JQU4 E0VKU6 A0A182NNA6 A0A182QEM8 A0A182P610 A0A182FLU3 A0A182R2R9 A0A182WAR8 A0A2R7VWY3 A0A2P8XP32 A0A1B6FD66 A0A0A9Z548 A0A069DTY5 U5EYM4 A0A1B6H9K6 J9JV79 A0A1L8DLT4 A0A2S2NYZ1 A0A023F432 A0A0V0GBQ9 A0A224XMP1 A0A1I8Q213 A0A2J7QAM8 A0A2R7VRD3 A0A2H8TDV2 A0A2M4BKU8 A0A1B6D3C8 A0A336LEX5 A0A0P4VS43 A0A2M4BKV1 T1HPM5 A0A067QJX1 A0A336N0T3 A0A1I8M387 A0A0L0BN98 A0A034VVS6 A0A0K8WFM1 W8BTG0 A0A1A9V7X1 A0A1B0FMC9 A0A1A9Z6I3 B4N996 B3P1T7 A0A0L7L113 A0A1B0FDM0 A0A1S4EJS9 B4HKI0 B4QXA1 A0A1A9V0D2 A0A023GLA1 G3MNK3 B3M1Z4 L7MAC3 A0A1E1XCE3
A0A194R6R5 D2A510 A0A0T6AU23 A0A1W4WYZ7 A0A182G740 A0A182JVE7 B0W646 Q16V55 A0A1S4FN34 U4UQY5 A0A084VZT7 A0A182JCS5 A0A182TTQ9 Q7PMU2 A0A182LI88 A0A0L7R0R3 N6TSY6 A0A2M4AS08 A0A182TC88 A0A087ZYB6 K7J6P0 A0A2A3EED5 A0A232EDW6 A0A0J7KVB9 A0A1Q3F8N9 A0A154PHX3 A0A151IHH8 A0A0N0BH30 E2BWP8 F4W7E2 A0A151JTC6 A0A182YI49 A0A158NPV2 E9IC81 A0A195DTR1 A0A151X8N3 A0A195ATG3 A0A026WTZ5 A0A182MIN6 E2A3S3 W5JQU4 E0VKU6 A0A182NNA6 A0A182QEM8 A0A182P610 A0A182FLU3 A0A182R2R9 A0A182WAR8 A0A2R7VWY3 A0A2P8XP32 A0A1B6FD66 A0A0A9Z548 A0A069DTY5 U5EYM4 A0A1B6H9K6 J9JV79 A0A1L8DLT4 A0A2S2NYZ1 A0A023F432 A0A0V0GBQ9 A0A224XMP1 A0A1I8Q213 A0A2J7QAM8 A0A2R7VRD3 A0A2H8TDV2 A0A2M4BKU8 A0A1B6D3C8 A0A336LEX5 A0A0P4VS43 A0A2M4BKV1 T1HPM5 A0A067QJX1 A0A336N0T3 A0A1I8M387 A0A0L0BN98 A0A034VVS6 A0A0K8WFM1 W8BTG0 A0A1A9V7X1 A0A1B0FMC9 A0A1A9Z6I3 B4N996 B3P1T7 A0A0L7L113 A0A1B0FDM0 A0A1S4EJS9 B4HKI0 B4QXA1 A0A1A9V0D2 A0A023GLA1 G3MNK3 B3M1Z4 L7MAC3 A0A1E1XCE3
PDB
6Q8I
E-value=2.19756e-52,
Score=521
Ontologies
KEGG
GO
PANTHER
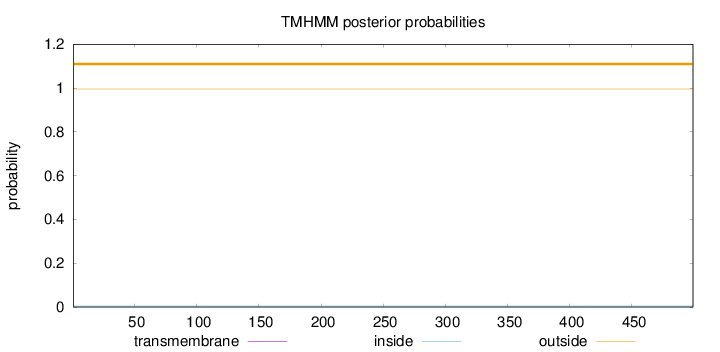
Topology
Length:
499
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00435
outside
1 - 499
Population Genetic Test Statistics
Pi
285.977527
Theta
200.64052
Tajima's D
1.213284
CLR
0.02078
CSRT
0.71906404679766
Interpretation
Uncertain
