Gene
KWMTBOMO09673
Pre Gene Modal
BGIBMGA012875
Annotation
PREDICTED:_protein_unc-119_homolog_[Papilio_polytes]
Full name
Protein unc-119 homolog
Location in the cell
Cytoplasmic Reliability : 2.608
Sequence
CDS
ATGAGCCTTGTCGGAAAAAAATGTGAATCGATATCTTTACACGAATCAATTAAAGACGATGTGATTTCCAATCAAGTAACTCCTGAAGATGTATTGAGATTGGAGAGAATTACAGATACATACCTCTGCAGCCCTGAAGCTAATGTATACGACATCGATTTTACGAGATTTAAAATTCGAGATTTAGAAACTGGAACGGTGTTATTTGAAATAGCTAAACCTCCGACTGATTTCGGCGTCGATAATGATTCTTTGGAAGATTCCACAGAGGAGCCCTTGGATCCAAATGCGGGGAGATTTGTTCGTTATCAATTCACACCACAATTTCTGAAGTTGAAAACTGTTGGCGCTACAGTGGAATTCACAGTTGGGGCTCGTCCAGTTAACCATTTCAGAATGATTGAGAAGCACTTCTTCAGGGATACACTACTGAAAACTTTTGATTTTGAGTTTGGTTTCTGTATTCCATTCTCAAGAAACACTTGCGAGCACATTTATGAGTTTCCAACGTTGCCCCCTGACTTAGTTGAGGAAATGATTGCTGCTCCTTTCGAGACATGTTCTGACAGTTTTTACTTCGTGGACAACCAATTGGTGATGCATAACAAAGCAGATTATGCTTATAATGGAGGTATCAACAATTAA
Protein
MSLVGKKCESISLHESIKDDVISNQVTPEDVLRLERITDTYLCSPEANVYDIDFTRFKIRDLETGTVLFEIAKPPTDFGVDNDSLEDSTEEPLDPNAGRFVRYQFTPQFLKLKTVGATVEFTVGARPVNHFRMIEKHFFRDTLLKTFDFEFGFCIPFSRNTCEHIYEFPTLPPDLVEEMIAAPFETCSDSFYFVDNQLVMHNKADYAYNGGINN
Summary
Description
Myristoyl-binding protein that acts as a cargo adapter: specifically binds the myristoyl moiety of a subset of N-terminally myristoylated proteins and is required for their localization.
Similarity
Belongs to the PDE6D/unc-119 family.
Keywords
Complete proteome
Lipid-binding
Protein transport
Reference proteome
Transport
Feature
chain Protein unc-119 homolog
Uniprot
H9JTL3
A0A437B8L0
A0A1E1VZL8
A0A2H1VDV4
A0A2A4K2P1
A0A194Q660
+ More
A0A212FBR3 S4P734 A0A0L7L336 A0A194R182 A0A1Y1NFE6 D2A4S1 A0A0T6AVT6 A0A1I8NII5 A0A1W4XSZ7 A0A293MTA1 T1JIL9 A0A2J7Q6V6 N6TZY9 J3JT90 A0A131XFW6 A0A067QGG3 B4MEN5 A0A2A3EG81 A0A088ANQ4 A0A131YGK0 A0A224Z645 L7M825 A0A310SNQ7 B4N1X0 A0A1B0FI38 A0A1A9XYD3 A0A1A9UUY1 A0A1B0ASW7 A0A1A9ZLQ3 A0A232ER15 A0A1E1XQC1 E0VJ90 A0A1E1X8R4 A0A1A9X034 A0A023GGM5 G3MPB8 B4JMV7 A0A1I8QD67 A0A1W4VJT6 A0A158NE03 A0A0M3QYX9 B3MQR6 B3NTP4 A0A0M8ZXL8 A0A1S4EV41 B4Q132 Q17PY4 A0A026W228 A0A195BPS4 B4L2C4 V5GXX2 A0A3B0JYD7 Q29GA9 A0A0L7R150 A0A1Q3F4D0 B4GVP5 B7PKQ8 A0A195ESF9 A0A226E5N3 E2BFE2 B0W0P6 E2AX70 A0A195C4V5 A0A195D810 A0A151X4G4 F4X1T1 A0A0A1X2J4 K1S270 A0A182R4B6 A0A443RLE2 C3ZL58 K7IRF0 A0A164PX86 Q0E5I1 Q540Y4 Q9XYQ2 Q0E5K0 Q0E5I2 Q0E5J1 A0A0P6BYN2 B4IHK8 A0A1B6D0H6 Q0E5J3 A0A0P5C1Q2 A0A0P5B5M0 E9HDJ9 A0A0P6AUT0 A0A034V0C4 A0A182U2A8 A0A182XKQ7 A0A182L4G1 A0A0K8W853 Q7PZ37 A0A3B3R0V4
A0A212FBR3 S4P734 A0A0L7L336 A0A194R182 A0A1Y1NFE6 D2A4S1 A0A0T6AVT6 A0A1I8NII5 A0A1W4XSZ7 A0A293MTA1 T1JIL9 A0A2J7Q6V6 N6TZY9 J3JT90 A0A131XFW6 A0A067QGG3 B4MEN5 A0A2A3EG81 A0A088ANQ4 A0A131YGK0 A0A224Z645 L7M825 A0A310SNQ7 B4N1X0 A0A1B0FI38 A0A1A9XYD3 A0A1A9UUY1 A0A1B0ASW7 A0A1A9ZLQ3 A0A232ER15 A0A1E1XQC1 E0VJ90 A0A1E1X8R4 A0A1A9X034 A0A023GGM5 G3MPB8 B4JMV7 A0A1I8QD67 A0A1W4VJT6 A0A158NE03 A0A0M3QYX9 B3MQR6 B3NTP4 A0A0M8ZXL8 A0A1S4EV41 B4Q132 Q17PY4 A0A026W228 A0A195BPS4 B4L2C4 V5GXX2 A0A3B0JYD7 Q29GA9 A0A0L7R150 A0A1Q3F4D0 B4GVP5 B7PKQ8 A0A195ESF9 A0A226E5N3 E2BFE2 B0W0P6 E2AX70 A0A195C4V5 A0A195D810 A0A151X4G4 F4X1T1 A0A0A1X2J4 K1S270 A0A182R4B6 A0A443RLE2 C3ZL58 K7IRF0 A0A164PX86 Q0E5I1 Q540Y4 Q9XYQ2 Q0E5K0 Q0E5I2 Q0E5J1 A0A0P6BYN2 B4IHK8 A0A1B6D0H6 Q0E5J3 A0A0P5C1Q2 A0A0P5B5M0 E9HDJ9 A0A0P6AUT0 A0A034V0C4 A0A182U2A8 A0A182XKQ7 A0A182L4G1 A0A0K8W853 Q7PZ37 A0A3B3R0V4
Pubmed
19121390
26354079
22118469
23622113
26227816
28004739
+ More
18362917 19820115 25315136 23537049 22516182 28049606 24845553 17994087 18057021 26830274 28797301 25576852 28648823 29209593 20566863 28503490 22216098 21347285 17550304 17510324 24508170 30249741 25765539 15632085 20798317 21719571 25830018 22992520 18563158 20075255 16868022 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 10858820 21292972 25348373 20966253 12364791 14747013 17210077 29240929
18362917 19820115 25315136 23537049 22516182 28049606 24845553 17994087 18057021 26830274 28797301 25576852 28648823 29209593 20566863 28503490 22216098 21347285 17550304 17510324 24508170 30249741 25765539 15632085 20798317 21719571 25830018 22992520 18563158 20075255 16868022 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 10858820 21292972 25348373 20966253 12364791 14747013 17210077 29240929
EMBL
BABH01003642
RSAL01000115
RVE46886.1
GDQN01010953
GDQN01007977
JAT80101.1
+ More
JAT83077.1 ODYU01002014 SOQ38997.1 NWSH01000261 PCG77912.1 KQ459582 KPI98895.1 AGBW02009293 OWR51158.1 GAIX01004774 JAA87786.1 JTDY01003327 KOB69731.1 KQ460878 KPJ11553.1 GEZM01003640 JAV96663.1 KQ971344 EFA05260.1 LJIG01022739 KRT78965.1 GFWV01019304 MAA44032.1 JH431806 NEVH01017450 PNF24313.1 APGK01054639 KB741251 ENN71837.1 BT126446 KB632429 AEE61410.1 ERL95481.1 GEFH01003339 JAP65242.1 KK853478 KDR07326.1 CH940664 EDW63010.1 KRF80870.1 KRF80871.1 KZ288254 PBC30783.1 GEDV01010188 JAP78369.1 GFPF01010626 MAA21772.1 GACK01005775 JAA59259.1 KQ759767 OAD62968.1 CH963925 EDW78359.1 CCAG010019914 JXJN01003024 NNAY01002670 OXU20810.1 GFAA01001936 JAU01499.1 DS235221 EEB13446.1 GFAC01003525 JAT95663.1 GBBM01002287 JAC33131.1 JO843719 AEO35336.1 CH916371 EDV92050.1 ADTU01013083 CP012528 ALC48399.1 CH902622 EDV34121.1 CH954180 EDV47457.1 KQ435833 KOX71663.1 CM000162 EDX02387.1 CH477188 EAT48810.1 KK107503 QOIP01000006 EZA49636.1 RLU22033.1 KQ976424 KYM88510.1 CH933810 EDW07785.1 GANP01009098 JAB75370.1 OUUW01000003 SPP78386.1 CH379064 EAL32201.1 KQ414668 KOC64569.1 GFDL01012666 JAV22379.1 CH479193 EDW26740.1 ABJB010284502 ABJB010451933 ABJB010508437 ABJB010512945 DS735207 EEC07180.1 KQ981993 KYN30824.1 LNIX01000006 OXA52982.1 GL448009 EFN85602.1 DS231818 EDS41348.1 GL443520 EFN61953.1 KQ978251 KYM95904.1 KQ981153 KYN09008.1 KQ982548 KYQ55313.1 GL888551 EGI59588.1 GBXI01008723 JAD05569.1 JH816368 EKC41401.1 NCKU01000305 NCKU01000300 RWS16075.1 RWS16090.1 GG666640 EEN46740.1 LRGB01002536 KZS07231.1 AM284485 AM284486 AM284488 AM284489 AM284490 AM284491 AM284492 AM284494 AM284487 CAK95967.1 CAK95968.1 CAK95969.1 CAK95970.1 CAK95971.1 CAK95972.1 CAK95973.1 CAK95975.1 CAK97611.2 AY118580 AE014298 AM284470 AM284471 AM284472 AM284473 AM284474 AM284476 AM284477 AM284478 AM284479 AM284480 AM284481 AM284483 AAM49949.1 AHN59449.1 CAK95952.1 CAK95953.1 CAK95954.1 CAK95955.1 CAK95956.1 CAK95958.1 CAK95959.1 CAK95960.1 CAK95961.1 CAK95962.1 CAK95963.1 CAK95965.1 AF119102 AM284475 CAK95957.1 AM284493 CAK95974.1 AM284484 CAK95966.1 GDIP01023286 JAM80429.1 CH480839 EDW49359.1 GEDC01018120 GEDC01015275 GEDC01009935 JAS19178.1 JAS22023.1 JAS27363.1 AM284482 CAK95964.1 GDIP01180694 JAJ42708.1 GDIP01204202 JAJ19200.1 GL732624 EFX70156.1 GDIP01024102 JAM79613.1 GAKP01023361 JAC35597.1 GDHF01005062 JAI47252.1 AAAB01008986 EAA00078.3
JAT83077.1 ODYU01002014 SOQ38997.1 NWSH01000261 PCG77912.1 KQ459582 KPI98895.1 AGBW02009293 OWR51158.1 GAIX01004774 JAA87786.1 JTDY01003327 KOB69731.1 KQ460878 KPJ11553.1 GEZM01003640 JAV96663.1 KQ971344 EFA05260.1 LJIG01022739 KRT78965.1 GFWV01019304 MAA44032.1 JH431806 NEVH01017450 PNF24313.1 APGK01054639 KB741251 ENN71837.1 BT126446 KB632429 AEE61410.1 ERL95481.1 GEFH01003339 JAP65242.1 KK853478 KDR07326.1 CH940664 EDW63010.1 KRF80870.1 KRF80871.1 KZ288254 PBC30783.1 GEDV01010188 JAP78369.1 GFPF01010626 MAA21772.1 GACK01005775 JAA59259.1 KQ759767 OAD62968.1 CH963925 EDW78359.1 CCAG010019914 JXJN01003024 NNAY01002670 OXU20810.1 GFAA01001936 JAU01499.1 DS235221 EEB13446.1 GFAC01003525 JAT95663.1 GBBM01002287 JAC33131.1 JO843719 AEO35336.1 CH916371 EDV92050.1 ADTU01013083 CP012528 ALC48399.1 CH902622 EDV34121.1 CH954180 EDV47457.1 KQ435833 KOX71663.1 CM000162 EDX02387.1 CH477188 EAT48810.1 KK107503 QOIP01000006 EZA49636.1 RLU22033.1 KQ976424 KYM88510.1 CH933810 EDW07785.1 GANP01009098 JAB75370.1 OUUW01000003 SPP78386.1 CH379064 EAL32201.1 KQ414668 KOC64569.1 GFDL01012666 JAV22379.1 CH479193 EDW26740.1 ABJB010284502 ABJB010451933 ABJB010508437 ABJB010512945 DS735207 EEC07180.1 KQ981993 KYN30824.1 LNIX01000006 OXA52982.1 GL448009 EFN85602.1 DS231818 EDS41348.1 GL443520 EFN61953.1 KQ978251 KYM95904.1 KQ981153 KYN09008.1 KQ982548 KYQ55313.1 GL888551 EGI59588.1 GBXI01008723 JAD05569.1 JH816368 EKC41401.1 NCKU01000305 NCKU01000300 RWS16075.1 RWS16090.1 GG666640 EEN46740.1 LRGB01002536 KZS07231.1 AM284485 AM284486 AM284488 AM284489 AM284490 AM284491 AM284492 AM284494 AM284487 CAK95967.1 CAK95968.1 CAK95969.1 CAK95970.1 CAK95971.1 CAK95972.1 CAK95973.1 CAK95975.1 CAK97611.2 AY118580 AE014298 AM284470 AM284471 AM284472 AM284473 AM284474 AM284476 AM284477 AM284478 AM284479 AM284480 AM284481 AM284483 AAM49949.1 AHN59449.1 CAK95952.1 CAK95953.1 CAK95954.1 CAK95955.1 CAK95956.1 CAK95958.1 CAK95959.1 CAK95960.1 CAK95961.1 CAK95962.1 CAK95963.1 CAK95965.1 AF119102 AM284475 CAK95957.1 AM284493 CAK95974.1 AM284484 CAK95966.1 GDIP01023286 JAM80429.1 CH480839 EDW49359.1 GEDC01018120 GEDC01015275 GEDC01009935 JAS19178.1 JAS22023.1 JAS27363.1 AM284482 CAK95964.1 GDIP01180694 JAJ42708.1 GDIP01204202 JAJ19200.1 GL732624 EFX70156.1 GDIP01024102 JAM79613.1 GAKP01023361 JAC35597.1 GDHF01005062 JAI47252.1 AAAB01008986 EAA00078.3
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000007151
UP000037510
+ More
UP000053240 UP000007266 UP000095301 UP000192223 UP000235965 UP000019118 UP000030742 UP000027135 UP000008792 UP000242457 UP000005203 UP000007798 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000215335 UP000009046 UP000091820 UP000001070 UP000095300 UP000192221 UP000005205 UP000092553 UP000007801 UP000008711 UP000053105 UP000002282 UP000008820 UP000053097 UP000279307 UP000078540 UP000009192 UP000268350 UP000001819 UP000053825 UP000008744 UP000001555 UP000078541 UP000198287 UP000008237 UP000002320 UP000000311 UP000078542 UP000078492 UP000075809 UP000007755 UP000005408 UP000075900 UP000285301 UP000001554 UP000002358 UP000076858 UP000000803 UP000001292 UP000000305 UP000075902 UP000076407 UP000075882 UP000007062 UP000261540
UP000053240 UP000007266 UP000095301 UP000192223 UP000235965 UP000019118 UP000030742 UP000027135 UP000008792 UP000242457 UP000005203 UP000007798 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000215335 UP000009046 UP000091820 UP000001070 UP000095300 UP000192221 UP000005205 UP000092553 UP000007801 UP000008711 UP000053105 UP000002282 UP000008820 UP000053097 UP000279307 UP000078540 UP000009192 UP000268350 UP000001819 UP000053825 UP000008744 UP000001555 UP000078541 UP000198287 UP000008237 UP000002320 UP000000311 UP000078542 UP000078492 UP000075809 UP000007755 UP000005408 UP000075900 UP000285301 UP000001554 UP000002358 UP000076858 UP000000803 UP000001292 UP000000305 UP000075902 UP000076407 UP000075882 UP000007062 UP000261540
Pfam
PF05351 GMP_PDE_delta
Interpro
SUPFAM
SSF81296
SSF81296
Gene 3D
ProteinModelPortal
H9JTL3
A0A437B8L0
A0A1E1VZL8
A0A2H1VDV4
A0A2A4K2P1
A0A194Q660
+ More
A0A212FBR3 S4P734 A0A0L7L336 A0A194R182 A0A1Y1NFE6 D2A4S1 A0A0T6AVT6 A0A1I8NII5 A0A1W4XSZ7 A0A293MTA1 T1JIL9 A0A2J7Q6V6 N6TZY9 J3JT90 A0A131XFW6 A0A067QGG3 B4MEN5 A0A2A3EG81 A0A088ANQ4 A0A131YGK0 A0A224Z645 L7M825 A0A310SNQ7 B4N1X0 A0A1B0FI38 A0A1A9XYD3 A0A1A9UUY1 A0A1B0ASW7 A0A1A9ZLQ3 A0A232ER15 A0A1E1XQC1 E0VJ90 A0A1E1X8R4 A0A1A9X034 A0A023GGM5 G3MPB8 B4JMV7 A0A1I8QD67 A0A1W4VJT6 A0A158NE03 A0A0M3QYX9 B3MQR6 B3NTP4 A0A0M8ZXL8 A0A1S4EV41 B4Q132 Q17PY4 A0A026W228 A0A195BPS4 B4L2C4 V5GXX2 A0A3B0JYD7 Q29GA9 A0A0L7R150 A0A1Q3F4D0 B4GVP5 B7PKQ8 A0A195ESF9 A0A226E5N3 E2BFE2 B0W0P6 E2AX70 A0A195C4V5 A0A195D810 A0A151X4G4 F4X1T1 A0A0A1X2J4 K1S270 A0A182R4B6 A0A443RLE2 C3ZL58 K7IRF0 A0A164PX86 Q0E5I1 Q540Y4 Q9XYQ2 Q0E5K0 Q0E5I2 Q0E5J1 A0A0P6BYN2 B4IHK8 A0A1B6D0H6 Q0E5J3 A0A0P5C1Q2 A0A0P5B5M0 E9HDJ9 A0A0P6AUT0 A0A034V0C4 A0A182U2A8 A0A182XKQ7 A0A182L4G1 A0A0K8W853 Q7PZ37 A0A3B3R0V4
A0A212FBR3 S4P734 A0A0L7L336 A0A194R182 A0A1Y1NFE6 D2A4S1 A0A0T6AVT6 A0A1I8NII5 A0A1W4XSZ7 A0A293MTA1 T1JIL9 A0A2J7Q6V6 N6TZY9 J3JT90 A0A131XFW6 A0A067QGG3 B4MEN5 A0A2A3EG81 A0A088ANQ4 A0A131YGK0 A0A224Z645 L7M825 A0A310SNQ7 B4N1X0 A0A1B0FI38 A0A1A9XYD3 A0A1A9UUY1 A0A1B0ASW7 A0A1A9ZLQ3 A0A232ER15 A0A1E1XQC1 E0VJ90 A0A1E1X8R4 A0A1A9X034 A0A023GGM5 G3MPB8 B4JMV7 A0A1I8QD67 A0A1W4VJT6 A0A158NE03 A0A0M3QYX9 B3MQR6 B3NTP4 A0A0M8ZXL8 A0A1S4EV41 B4Q132 Q17PY4 A0A026W228 A0A195BPS4 B4L2C4 V5GXX2 A0A3B0JYD7 Q29GA9 A0A0L7R150 A0A1Q3F4D0 B4GVP5 B7PKQ8 A0A195ESF9 A0A226E5N3 E2BFE2 B0W0P6 E2AX70 A0A195C4V5 A0A195D810 A0A151X4G4 F4X1T1 A0A0A1X2J4 K1S270 A0A182R4B6 A0A443RLE2 C3ZL58 K7IRF0 A0A164PX86 Q0E5I1 Q540Y4 Q9XYQ2 Q0E5K0 Q0E5I2 Q0E5J1 A0A0P6BYN2 B4IHK8 A0A1B6D0H6 Q0E5J3 A0A0P5C1Q2 A0A0P5B5M0 E9HDJ9 A0A0P6AUT0 A0A034V0C4 A0A182U2A8 A0A182XKQ7 A0A182L4G1 A0A0K8W853 Q7PZ37 A0A3B3R0V4
PDB
4GOK
E-value=3.69802e-68,
Score=652
Ontologies
PANTHER
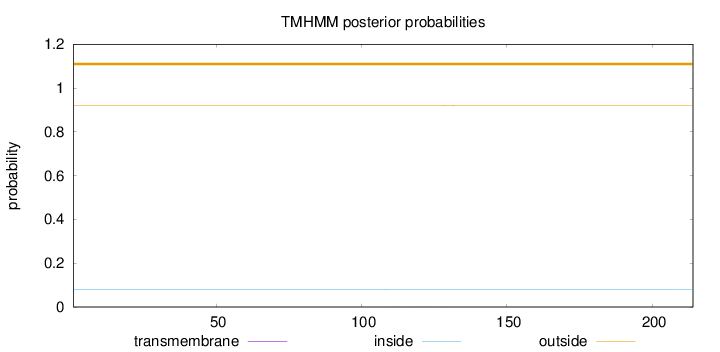
Topology
Length:
214
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00913999999999998
Exp number, first 60 AAs:
0
Total prob of N-in:
0.07917
outside
1 - 214
Population Genetic Test Statistics
Pi
250.761818
Theta
221.866781
Tajima's D
0.50494
CLR
0.535138
CSRT
0.514624268786561
Interpretation
Uncertain
