Gene
KWMTBOMO09672 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012960
Annotation
PREDICTED:_phosphatidylinositol_transfer_protein_alpha_isoform_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 4.062
Sequence
CDS
ATGATTATCAAAGAATACCGAGTCACACTTCCTCTAACAGTTGAAGAGTATCAAGTGGCCCAGTTATACTGCGTGGCGGAGGTCTCCAAAAATGAAACCGGCGGAGGTGAAGGCATAGAGGTCATAAAGAATGAACCGTTCAAAGATTATCCACTGCTCGGTGGAAAATACTCGTCTGGACAGTACACGTACAAGATCTACCATTTGGCGTCCAAAGTTCCGGCCTTCATCAGGCTACTTGCTCCGAAAGGATCTCTCGAAGTACACGAAGAAGCGTGGAACGCCTACCCGTACTGTAGAACAGTGCTGACGAATCCCGGTTACATGAAAGAAAACTTCGTTATATGTATAGAATCGCTGCATTTACCGGACGCCGGCGACCAATACAATGTCCACGAGCTGCCGCCTGAGAAACTAAAGACCCGCGAGGTGGTCCCCATAGACATTGCGAACGACCCCCTCCCGTCGGCCGACTACAAATCGGAGACCGACCCGACCAAGTTCAAGTCTACGAAGACCGGCCGCGGACCGCTCGTCGGGCCAAACTGGAAGAACGACGTGAAACCGGTCATGACGTGCTACAAGCTCGTCACGGCCGAGTTCAAATGGTTTGGTCTCCAGAATAAGGTAGAGAATGTCATCCAGAAATCGGAGCGACGATTGTTCACAATCTTTCACAGACAAGTGTTCTGTTCGATGGACGACTGGTACGGAATGACGATGGCCGACATCCGAGCTATCGAGGATCGCACCAAGGAGGAGTTGGAGAAGCTGCGGCGCGAGGGAGAGGTGCGCGGCATGCGGGCCGACAACGAGTAG
Protein
MIIKEYRVTLPLTVEEYQVAQLYCVAEVSKNETGGGEGIEVIKNEPFKDYPLLGGKYSSGQYTYKIYHLASKVPAFIRLLAPKGSLEVHEEAWNAYPYCRTVLTNPGYMKENFVICIESLHLPDAGDQYNVHELPPEKLKTREVVPIDIANDPLPSADYKSETDPTKFKSTKTGRGPLVGPNWKNDVKPVMTCYKLVTAEFKWFGLQNKVENVIQKSERRLFTIFHRQVFCSMDDWYGMTMADIRAIEDRTKEELEKLRREGEVRGMRADNE
Summary
Uniprot
A0A437B950
A0A2A4K0W6
A0A1E1WGR4
A0A1E1WIC1
S4PEZ6
A0A194R1H9
+ More
A0A194Q002 A0A212FBR1 A0A2M4A4G4 A0A0K8UKS6 A0A2M3Z1L6 A0A2M4CUM2 Q7PUN0 A0A182QLS2 A0A182KLY1 A0A1W4XLR0 A0A1I8MLN0 A0A1Y1KGU7 A0A336K9M3 V9IGA9 A0A088AUP1 A0A0J7L2C9 E2C414 J3JXZ3 W5J953 A0A182JLP1 D6WYS3 A0A1L8DYZ2 A0A158NBS6 A0A1L8DZ08 A0A1L8EH07 A0A182N4C2 Q17DK4 W8BHP2 A0A1W7R8B1 A0A0K8TN39 K7ISK7 U5EY28 E2A4U5 A0A0A1XLV7 T1PCP0 A0A0A9YWM3 A0A0K8VZX3 A0A034W637 A0A1Q3F455 D3TN38 A0A1J1IK49 A0A3L8DVI2 A0A2M4A4K6 A0A1I8PUA1 A0A1B0G7Y9 A0A232FH25 A0A1B6FGX0 A0A1D2MSE6 A0A2M4CTD5 F5HMB4 A0A1B6IDS7 B4NIT6 A0A0M8ZTK5 B3M2F0 B4QTF8 B0WRR8 B4PMT1 A0A1W4VXB7 Q9VDY8 B4KD22 B3P360 A0A2R7WUT4 A0A3B0K6U1 Q8T3L2 A0A1A9ZHM1 B4JUX7 A0A1B0BNL3 B4LZK9 A0A2A3ERT7 Q293P7 B4GLT5 A0A224XUC6 A0A1A9Y6N8 A0A1A9WJA5 A0A067R8Y0 A0A154PMU0 A0A069DYB7 A0A0P4W378 R4G5F6 A0A0Q9X9T5 A0A0L7QKW7 C4WTL9 A0A182GT93 A0A0R3NKD9 A0A182PQU4 A0A182TPU0 A0A182XBG2 A0A182I0A2 A0A2L2YP22 A0A0R1E580 A0A0Q9WF02 A0A0B4KGG5 A0A226DKW4 A0A2H8TU05
A0A194Q002 A0A212FBR1 A0A2M4A4G4 A0A0K8UKS6 A0A2M3Z1L6 A0A2M4CUM2 Q7PUN0 A0A182QLS2 A0A182KLY1 A0A1W4XLR0 A0A1I8MLN0 A0A1Y1KGU7 A0A336K9M3 V9IGA9 A0A088AUP1 A0A0J7L2C9 E2C414 J3JXZ3 W5J953 A0A182JLP1 D6WYS3 A0A1L8DYZ2 A0A158NBS6 A0A1L8DZ08 A0A1L8EH07 A0A182N4C2 Q17DK4 W8BHP2 A0A1W7R8B1 A0A0K8TN39 K7ISK7 U5EY28 E2A4U5 A0A0A1XLV7 T1PCP0 A0A0A9YWM3 A0A0K8VZX3 A0A034W637 A0A1Q3F455 D3TN38 A0A1J1IK49 A0A3L8DVI2 A0A2M4A4K6 A0A1I8PUA1 A0A1B0G7Y9 A0A232FH25 A0A1B6FGX0 A0A1D2MSE6 A0A2M4CTD5 F5HMB4 A0A1B6IDS7 B4NIT6 A0A0M8ZTK5 B3M2F0 B4QTF8 B0WRR8 B4PMT1 A0A1W4VXB7 Q9VDY8 B4KD22 B3P360 A0A2R7WUT4 A0A3B0K6U1 Q8T3L2 A0A1A9ZHM1 B4JUX7 A0A1B0BNL3 B4LZK9 A0A2A3ERT7 Q293P7 B4GLT5 A0A224XUC6 A0A1A9Y6N8 A0A1A9WJA5 A0A067R8Y0 A0A154PMU0 A0A069DYB7 A0A0P4W378 R4G5F6 A0A0Q9X9T5 A0A0L7QKW7 C4WTL9 A0A182GT93 A0A0R3NKD9 A0A182PQU4 A0A182TPU0 A0A182XBG2 A0A182I0A2 A0A2L2YP22 A0A0R1E580 A0A0Q9WF02 A0A0B4KGG5 A0A226DKW4 A0A2H8TU05
Pubmed
23622113
26354079
22118469
12364791
14747013
17210077
+ More
20966253 25315136 28004739 20798317 22516182 20920257 23761445 18362917 19820115 21347285 17510324 24495485 26369729 20075255 25830018 25401762 26823975 25348373 20353571 30249741 28648823 27289101 17994087 18057021 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 24845553 26334808 27129103 26483478 26561354
20966253 25315136 28004739 20798317 22516182 20920257 23761445 18362917 19820115 21347285 17510324 24495485 26369729 20075255 25830018 25401762 26823975 25348373 20353571 30249741 28648823 27289101 17994087 18057021 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 24845553 26334808 27129103 26483478 26561354
EMBL
RSAL01000115
RVE46885.1
NWSH01000261
PCG77911.1
GDQN01004885
JAT86169.1
+ More
GDQN01004291 JAT86763.1 GAIX01004272 JAA88288.1 KQ460878 KPJ11552.1 KQ459582 KPI98896.1 AGBW02009293 OWR51157.1 GGFK01002383 MBW35704.1 GDHF01025176 JAI27138.1 GGFM01001659 MBW22410.1 GGFL01004360 MBW68538.1 AAAB01008987 EAA01271.5 AXCN02000031 AXCN02000032 GEZM01084713 JAV60709.1 UFQS01000197 UFQT01000197 SSX01191.1 SSX21571.1 JR042934 AEY59692.1 LBMM01001173 KMQ96783.1 KMQ96784.1 GL452364 EFN77400.1 BT128115 AEE63076.1 ADMH02002026 ETN59908.1 KQ971357 EFA08400.1 GFDF01002394 JAV11690.1 ADTU01011295 GFDF01002374 JAV11710.1 GFDG01000828 JAV17971.1 CH477293 EAT44497.1 GAMC01017331 GAMC01017330 JAB89224.1 GEHC01000257 JAV47388.1 GDAI01001834 JAI15769.1 GANO01000581 JAB59290.1 GL436748 EFN71539.1 GBXI01002291 JAD12001.1 KA646449 AFP61078.1 GBHO01006182 GBRD01007473 GDHC01004718 JAG37422.1 JAG58348.1 JAQ13911.1 GDHF01007887 JAI44427.1 GAKP01009739 GAKP01009738 JAC49213.1 GFDL01012722 JAV22323.1 EZ422840 ADD19116.1 CVRI01000048 CRK98825.1 QOIP01000004 RLU24233.1 GGFK01002360 MBW35681.1 CCAG010010579 CCAG010010580 NNAY01000198 OXU30066.1 GECZ01020556 JAS49213.1 LJIJ01000593 ODM96017.1 GGFL01004361 MBW68539.1 EGK97436.1 GECU01029243 GECU01022656 JAS78463.1 JAS85050.1 CH964272 EDW83800.1 KQ435883 KOX69696.1 CH902617 EDV43403.1 KPU80265.1 CM000364 EDX12360.1 DS232059 EDS33477.1 CM000160 EDW96001.1 KRK02836.1 AE014297 AF246993 BT083455 AAF55650.1 AAF61273.1 ACR16680.1 AHN57401.1 CH933806 EDW13792.1 CH954181 EDV48512.1 KK855626 PTY23327.1 OUUW01000008 SPP83780.1 AY094957 AAM11310.1 CH916374 EDV91297.1 JXJN01017442 JXJN01017443 CH940650 EDW67148.1 KZ288192 PBC34224.1 CM000070 EAL29167.2 CH479185 EDW38509.1 GFTR01004396 JAW12030.1 KK852619 KDR20111.1 KQ434954 KZC12530.1 GBGD01002510 JAC86379.1 GDKW01000664 JAI55931.1 ACPB03014848 GAHY01000610 JAA76900.1 KRG00643.1 KRG00644.1 KQ414934 KOC59262.1 AK340674 BAH71239.1 JXUM01086739 JXUM01086740 JXUM01086741 JXUM01086742 JXUM01086743 KQ563588 KXJ73626.1 KRT01332.1 APCN01002057 IAAA01040882 LAA09888.1 KRK02833.1 KRK02834.1 KRK02835.1 KRF83133.1 AGB96106.1 LNIX01000017 OXA45628.1 GFXV01005929 MBW17734.1
GDQN01004291 JAT86763.1 GAIX01004272 JAA88288.1 KQ460878 KPJ11552.1 KQ459582 KPI98896.1 AGBW02009293 OWR51157.1 GGFK01002383 MBW35704.1 GDHF01025176 JAI27138.1 GGFM01001659 MBW22410.1 GGFL01004360 MBW68538.1 AAAB01008987 EAA01271.5 AXCN02000031 AXCN02000032 GEZM01084713 JAV60709.1 UFQS01000197 UFQT01000197 SSX01191.1 SSX21571.1 JR042934 AEY59692.1 LBMM01001173 KMQ96783.1 KMQ96784.1 GL452364 EFN77400.1 BT128115 AEE63076.1 ADMH02002026 ETN59908.1 KQ971357 EFA08400.1 GFDF01002394 JAV11690.1 ADTU01011295 GFDF01002374 JAV11710.1 GFDG01000828 JAV17971.1 CH477293 EAT44497.1 GAMC01017331 GAMC01017330 JAB89224.1 GEHC01000257 JAV47388.1 GDAI01001834 JAI15769.1 GANO01000581 JAB59290.1 GL436748 EFN71539.1 GBXI01002291 JAD12001.1 KA646449 AFP61078.1 GBHO01006182 GBRD01007473 GDHC01004718 JAG37422.1 JAG58348.1 JAQ13911.1 GDHF01007887 JAI44427.1 GAKP01009739 GAKP01009738 JAC49213.1 GFDL01012722 JAV22323.1 EZ422840 ADD19116.1 CVRI01000048 CRK98825.1 QOIP01000004 RLU24233.1 GGFK01002360 MBW35681.1 CCAG010010579 CCAG010010580 NNAY01000198 OXU30066.1 GECZ01020556 JAS49213.1 LJIJ01000593 ODM96017.1 GGFL01004361 MBW68539.1 EGK97436.1 GECU01029243 GECU01022656 JAS78463.1 JAS85050.1 CH964272 EDW83800.1 KQ435883 KOX69696.1 CH902617 EDV43403.1 KPU80265.1 CM000364 EDX12360.1 DS232059 EDS33477.1 CM000160 EDW96001.1 KRK02836.1 AE014297 AF246993 BT083455 AAF55650.1 AAF61273.1 ACR16680.1 AHN57401.1 CH933806 EDW13792.1 CH954181 EDV48512.1 KK855626 PTY23327.1 OUUW01000008 SPP83780.1 AY094957 AAM11310.1 CH916374 EDV91297.1 JXJN01017442 JXJN01017443 CH940650 EDW67148.1 KZ288192 PBC34224.1 CM000070 EAL29167.2 CH479185 EDW38509.1 GFTR01004396 JAW12030.1 KK852619 KDR20111.1 KQ434954 KZC12530.1 GBGD01002510 JAC86379.1 GDKW01000664 JAI55931.1 ACPB03014848 GAHY01000610 JAA76900.1 KRG00643.1 KRG00644.1 KQ414934 KOC59262.1 AK340674 BAH71239.1 JXUM01086739 JXUM01086740 JXUM01086741 JXUM01086742 JXUM01086743 KQ563588 KXJ73626.1 KRT01332.1 APCN01002057 IAAA01040882 LAA09888.1 KRK02833.1 KRK02834.1 KRK02835.1 KRF83133.1 AGB96106.1 LNIX01000017 OXA45628.1 GFXV01005929 MBW17734.1
Proteomes
UP000283053
UP000218220
UP000053240
UP000053268
UP000007151
UP000007062
+ More
UP000075886 UP000075882 UP000192223 UP000095301 UP000005203 UP000036403 UP000008237 UP000000673 UP000075880 UP000007266 UP000005205 UP000075884 UP000008820 UP000002358 UP000000311 UP000183832 UP000279307 UP000095300 UP000092444 UP000215335 UP000094527 UP000007798 UP000053105 UP000007801 UP000000304 UP000002320 UP000002282 UP000192221 UP000000803 UP000009192 UP000008711 UP000268350 UP000092445 UP000001070 UP000092460 UP000008792 UP000242457 UP000001819 UP000008744 UP000092443 UP000091820 UP000027135 UP000076502 UP000015103 UP000053825 UP000069940 UP000249989 UP000075885 UP000075902 UP000076407 UP000075840 UP000198287
UP000075886 UP000075882 UP000192223 UP000095301 UP000005203 UP000036403 UP000008237 UP000000673 UP000075880 UP000007266 UP000005205 UP000075884 UP000008820 UP000002358 UP000000311 UP000183832 UP000279307 UP000095300 UP000092444 UP000215335 UP000094527 UP000007798 UP000053105 UP000007801 UP000000304 UP000002320 UP000002282 UP000192221 UP000000803 UP000009192 UP000008711 UP000268350 UP000092445 UP000001070 UP000092460 UP000008792 UP000242457 UP000001819 UP000008744 UP000092443 UP000091820 UP000027135 UP000076502 UP000015103 UP000053825 UP000069940 UP000249989 UP000075885 UP000075902 UP000076407 UP000075840 UP000198287
Pfam
PF02121 IP_trans
Gene 3D
ProteinModelPortal
A0A437B950
A0A2A4K0W6
A0A1E1WGR4
A0A1E1WIC1
S4PEZ6
A0A194R1H9
+ More
A0A194Q002 A0A212FBR1 A0A2M4A4G4 A0A0K8UKS6 A0A2M3Z1L6 A0A2M4CUM2 Q7PUN0 A0A182QLS2 A0A182KLY1 A0A1W4XLR0 A0A1I8MLN0 A0A1Y1KGU7 A0A336K9M3 V9IGA9 A0A088AUP1 A0A0J7L2C9 E2C414 J3JXZ3 W5J953 A0A182JLP1 D6WYS3 A0A1L8DYZ2 A0A158NBS6 A0A1L8DZ08 A0A1L8EH07 A0A182N4C2 Q17DK4 W8BHP2 A0A1W7R8B1 A0A0K8TN39 K7ISK7 U5EY28 E2A4U5 A0A0A1XLV7 T1PCP0 A0A0A9YWM3 A0A0K8VZX3 A0A034W637 A0A1Q3F455 D3TN38 A0A1J1IK49 A0A3L8DVI2 A0A2M4A4K6 A0A1I8PUA1 A0A1B0G7Y9 A0A232FH25 A0A1B6FGX0 A0A1D2MSE6 A0A2M4CTD5 F5HMB4 A0A1B6IDS7 B4NIT6 A0A0M8ZTK5 B3M2F0 B4QTF8 B0WRR8 B4PMT1 A0A1W4VXB7 Q9VDY8 B4KD22 B3P360 A0A2R7WUT4 A0A3B0K6U1 Q8T3L2 A0A1A9ZHM1 B4JUX7 A0A1B0BNL3 B4LZK9 A0A2A3ERT7 Q293P7 B4GLT5 A0A224XUC6 A0A1A9Y6N8 A0A1A9WJA5 A0A067R8Y0 A0A154PMU0 A0A069DYB7 A0A0P4W378 R4G5F6 A0A0Q9X9T5 A0A0L7QKW7 C4WTL9 A0A182GT93 A0A0R3NKD9 A0A182PQU4 A0A182TPU0 A0A182XBG2 A0A182I0A2 A0A2L2YP22 A0A0R1E580 A0A0Q9WF02 A0A0B4KGG5 A0A226DKW4 A0A2H8TU05
A0A194Q002 A0A212FBR1 A0A2M4A4G4 A0A0K8UKS6 A0A2M3Z1L6 A0A2M4CUM2 Q7PUN0 A0A182QLS2 A0A182KLY1 A0A1W4XLR0 A0A1I8MLN0 A0A1Y1KGU7 A0A336K9M3 V9IGA9 A0A088AUP1 A0A0J7L2C9 E2C414 J3JXZ3 W5J953 A0A182JLP1 D6WYS3 A0A1L8DYZ2 A0A158NBS6 A0A1L8DZ08 A0A1L8EH07 A0A182N4C2 Q17DK4 W8BHP2 A0A1W7R8B1 A0A0K8TN39 K7ISK7 U5EY28 E2A4U5 A0A0A1XLV7 T1PCP0 A0A0A9YWM3 A0A0K8VZX3 A0A034W637 A0A1Q3F455 D3TN38 A0A1J1IK49 A0A3L8DVI2 A0A2M4A4K6 A0A1I8PUA1 A0A1B0G7Y9 A0A232FH25 A0A1B6FGX0 A0A1D2MSE6 A0A2M4CTD5 F5HMB4 A0A1B6IDS7 B4NIT6 A0A0M8ZTK5 B3M2F0 B4QTF8 B0WRR8 B4PMT1 A0A1W4VXB7 Q9VDY8 B4KD22 B3P360 A0A2R7WUT4 A0A3B0K6U1 Q8T3L2 A0A1A9ZHM1 B4JUX7 A0A1B0BNL3 B4LZK9 A0A2A3ERT7 Q293P7 B4GLT5 A0A224XUC6 A0A1A9Y6N8 A0A1A9WJA5 A0A067R8Y0 A0A154PMU0 A0A069DYB7 A0A0P4W378 R4G5F6 A0A0Q9X9T5 A0A0L7QKW7 C4WTL9 A0A182GT93 A0A0R3NKD9 A0A182PQU4 A0A182TPU0 A0A182XBG2 A0A182I0A2 A0A2L2YP22 A0A0R1E580 A0A0Q9WF02 A0A0B4KGG5 A0A226DKW4 A0A2H8TU05
PDB
1UW5
E-value=3.53528e-83,
Score=783
Ontologies
GO
GO:0005548
GO:0031210
GO:0005737
GO:0008526
GO:0035091
GO:0008525
GO:0031965
GO:0005794
GO:0031566
GO:0000915
GO:0007111
GO:0000916
GO:0032154
GO:0007110
GO:0048137
GO:0007112
GO:0000212
GO:0016006
GO:0070732
GO:0005886
GO:0033206
GO:0005622
GO:0006810
GO:0016491
GO:0006508
GO:0016787
GO:0003723
GO:0006730
GO:0009258
GO:0016155
GO:0003824
GO:0015914
PANTHER
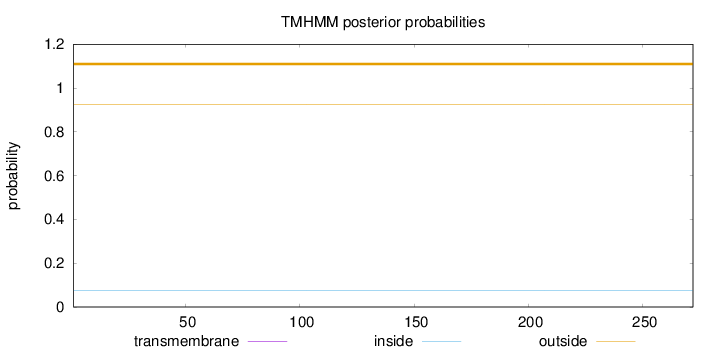
Topology
Length:
272
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00133
Exp number, first 60 AAs:
0.0001
Total prob of N-in:
0.07678
outside
1 - 272
Population Genetic Test Statistics
Pi
273.705645
Theta
174.785726
Tajima's D
1.635087
CLR
0.039697
CSRT
0.814309284535773
Interpretation
Uncertain
