Gene
KWMTBOMO09670
Annotation
PREDICTED:_dnaJ_(Hsp40)_homolog_11_isoform_X1_[Bombyx_mori]
Full name
S-adenosylmethionine sensor upstream of mTORC1
Alternative Name
Probable methyltransferase BMT2 homolog
Location in the cell
Mitochondrial Reliability : 1.397 Nuclear Reliability : 1.401
Sequence
CDS
ATGCTCCGTCAAAGAAACAATATTTTAGTGACAACTTTTTATAGACAATTTACTACCTCACCTCAATTTTGTGCCAGCCACTATGATGTGCTTGGAGTCACACCAAAAGCGACCCAGAACGATATAAAAACGGCTTACTATAAACTTTCCAAAGTACACCACCCGGACAAATCTAAGGATGAAGCATCTATCAAAAAGTTTCGAGCAATAACAGAAGCATATGAGGTACTTGGGAATATTCAACTTAAAAAAATGTATGATAGAGGTCTCTTAGTTGGTAGAGAGAATACTTCTCGTATGGAATACAAACCAGAACCAGAACCTACAGATCCTACCCTAAAGTTTTACAAGTCCCATGCAACTCGCAATATCACACCCACCATGGACGGGAAAGTTCCAATTTATGACTTTGATAGTTGGTCCAAACACCATTATTCAGATGTATTTGCAAAGCAGAAATATGATAAGGAAATGGTACGAAACTCACAAGAAAAACAACAGAAAATATTAGAATCTACTAAGCAAGAAGGAATTATTTGGCTTGTTATAATCATAGGTGGATTGTTTATTGCGGTGTTACTTAATGGACTCGATGATTATGATCAAAATCAGTTACAAAAGAATAATATAGAAAAAAAATGA
Protein
MLRQRNNILVTTFYRQFTTSPQFCASHYDVLGVTPKATQNDIKTAYYKLSKVHHPDKSKDEASIKKFRAITEAYEVLGNIQLKKMYDRGLLVGRENTSRMEYKPEPEPTDPTLKFYKSHATRNITPTMDGKVPIYDFDSWSKHHYSDVFAKQKYDKEMVRNSQEKQQKILESTKQEGIIWLVIIIGGLFIAVLLNGLDDYDQNQLQKNNIEKK
Summary
Description
S-adenosyl-L-methionine-binding protein that acts as an inhibitor of mTORC1 signaling. Acts as a sensor of S-adenosyl-L-methionine to signal methionine sufficiency to mTORC1. Probably also acts as a S-adenosyl-L-methionine-dependent methyltransferase.
Similarity
Belongs to the BMT2 family.
Uniprot
H6VTQ1
C7AQZ9
A0A194R200
A0A194Q1M8
S4PWU7
A0A437B8P7
+ More
A0A212FBN5 A0A2H1W555 A0A2A4JE72 A0A2A4JD33 A0A0L7KL18 A0A0K8TN67 B0W3K6 A0A1Q3FH55 A0A182K4L5 A0A182QQW9 A0A182F942 A0A182PKG2 A0A084W4Y9 A0A2M4BYC0 A0A182T1S0 A0A1D6TIB9 A0A182UPK9 A0A182TSS3 A0A182LAY8 Q5TVD0 A0A182I908 A0A182YP33 A0A182WVC7 W5JB29 A0A067R0M0 T1DM52 A0A2M3ZJN0 A0A182N4K7 A0A182VY17 A0A182M7X9 A0A1I8MRF4 A0A1I8N2P0 A0A1I8MT85 J3JZ42 A0A1I8PFR0 A0A1Y1M290 A0A0T6B8C2 A0A423S9T9 A0A3R7LYD5 Q295I0 B4GDV0 D2A3L8 B4JUS2 A0A336K4I6 A0A2J7PRC0 A0A1A9W8C0 A0A0P4W746 A0A1A9VFM1 A0A0L0C784 A0A1A9XWB1 N6SUV0 A0A1B0FCP7 T1HR47 U4UQX3 A0A1B0A375 B4NJ37 A0A1L8DTW7 A0A1L8DTP6 A0A1L8DU93 A0A0P4VM14 A0A1B6HVP4 A0A1B6FYQ9 E9FYC9 A0A1S3CYK0
A0A212FBN5 A0A2H1W555 A0A2A4JE72 A0A2A4JD33 A0A0L7KL18 A0A0K8TN67 B0W3K6 A0A1Q3FH55 A0A182K4L5 A0A182QQW9 A0A182F942 A0A182PKG2 A0A084W4Y9 A0A2M4BYC0 A0A182T1S0 A0A1D6TIB9 A0A182UPK9 A0A182TSS3 A0A182LAY8 Q5TVD0 A0A182I908 A0A182YP33 A0A182WVC7 W5JB29 A0A067R0M0 T1DM52 A0A2M3ZJN0 A0A182N4K7 A0A182VY17 A0A182M7X9 A0A1I8MRF4 A0A1I8N2P0 A0A1I8MT85 J3JZ42 A0A1I8PFR0 A0A1Y1M290 A0A0T6B8C2 A0A423S9T9 A0A3R7LYD5 Q295I0 B4GDV0 D2A3L8 B4JUS2 A0A336K4I6 A0A2J7PRC0 A0A1A9W8C0 A0A0P4W746 A0A1A9VFM1 A0A0L0C784 A0A1A9XWB1 N6SUV0 A0A1B0FCP7 T1HR47 U4UQX3 A0A1B0A375 B4NJ37 A0A1L8DTW7 A0A1L8DTP6 A0A1L8DU93 A0A0P4VM14 A0A1B6HVP4 A0A1B6FYQ9 E9FYC9 A0A1S3CYK0
Pubmed
EMBL
JN872886
AFC01225.1
FJ592084
ACT34045.1
KQ460878
KPJ11549.1
+ More
KQ459582 KPI98899.1 GAIX01006168 JAA86392.1 RSAL01000115 RVE46882.1 AGBW02009293 OWR51154.1 ODYU01006408 SOQ48225.1 NWSH01001822 PCG70008.1 PCG70007.1 JTDY01009381 KOB63589.1 GDAI01002233 JAI15370.1 DS231832 EDS31792.1 GFDL01008183 JAV26862.1 AXCN02000044 ATLV01020424 KE525302 KFB45283.1 GGFJ01008871 MBW58012.1 AAAB01008844 EAA05983.3 APCN01003147 ADMH02001893 ETN60613.1 KK852802 KDR16290.1 GAMD01000086 JAB01505.1 GGFM01007934 MBW28685.1 AXCM01004998 BT128523 AEE63480.1 GEZM01042299 GEZM01042298 JAV79954.1 LJIG01009223 KRT83509.1 QCYY01004455 ROT60971.1 QCYY01003696 ROT62401.1 CM000070 EAL28732.3 CH479182 EDW34611.1 KQ971348 EFA05522.1 CH916374 EDV91242.1 UFQS01000022 UFQT01000022 SSW97613.1 SSX17999.1 NEVH01022362 PNF18883.1 GDRN01084928 JAI61421.1 JRES01000819 KNC28130.1 APGK01055728 KB741269 ENN71469.1 CCAG010010183 ACPB03009189 KB632330 ERL92556.1 CH964272 EDW83830.1 GFDF01004257 JAV09827.1 GFDF01004258 JAV09826.1 GFDF01004259 JAV09825.1 GDKW01003119 JAI53476.1 GECU01028958 JAS78748.1 GECZ01014457 JAS55312.1 GL732527 EFX87793.1
KQ459582 KPI98899.1 GAIX01006168 JAA86392.1 RSAL01000115 RVE46882.1 AGBW02009293 OWR51154.1 ODYU01006408 SOQ48225.1 NWSH01001822 PCG70008.1 PCG70007.1 JTDY01009381 KOB63589.1 GDAI01002233 JAI15370.1 DS231832 EDS31792.1 GFDL01008183 JAV26862.1 AXCN02000044 ATLV01020424 KE525302 KFB45283.1 GGFJ01008871 MBW58012.1 AAAB01008844 EAA05983.3 APCN01003147 ADMH02001893 ETN60613.1 KK852802 KDR16290.1 GAMD01000086 JAB01505.1 GGFM01007934 MBW28685.1 AXCM01004998 BT128523 AEE63480.1 GEZM01042299 GEZM01042298 JAV79954.1 LJIG01009223 KRT83509.1 QCYY01004455 ROT60971.1 QCYY01003696 ROT62401.1 CM000070 EAL28732.3 CH479182 EDW34611.1 KQ971348 EFA05522.1 CH916374 EDV91242.1 UFQS01000022 UFQT01000022 SSW97613.1 SSX17999.1 NEVH01022362 PNF18883.1 GDRN01084928 JAI61421.1 JRES01000819 KNC28130.1 APGK01055728 KB741269 ENN71469.1 CCAG010010183 ACPB03009189 KB632330 ERL92556.1 CH964272 EDW83830.1 GFDF01004257 JAV09827.1 GFDF01004258 JAV09826.1 GFDF01004259 JAV09825.1 GDKW01003119 JAI53476.1 GECU01028958 JAS78748.1 GECZ01014457 JAS55312.1 GL732527 EFX87793.1
Proteomes
UP000053240
UP000053268
UP000283053
UP000007151
UP000218220
UP000037510
+ More
UP000002320 UP000075881 UP000075886 UP000069272 UP000075885 UP000030765 UP000075901 UP000076407 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840 UP000076408 UP000000673 UP000027135 UP000075884 UP000075920 UP000075883 UP000095301 UP000095300 UP000283509 UP000001819 UP000008744 UP000007266 UP000001070 UP000235965 UP000091820 UP000078200 UP000037069 UP000092443 UP000019118 UP000092444 UP000015103 UP000030742 UP000092445 UP000007798 UP000000305 UP000079169
UP000002320 UP000075881 UP000075886 UP000069272 UP000075885 UP000030765 UP000075901 UP000076407 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840 UP000076408 UP000000673 UP000027135 UP000075884 UP000075920 UP000075883 UP000095301 UP000095300 UP000283509 UP000001819 UP000008744 UP000007266 UP000001070 UP000235965 UP000091820 UP000078200 UP000037069 UP000092443 UP000019118 UP000092444 UP000015103 UP000030742 UP000092445 UP000007798 UP000000305 UP000079169
Interpro
Gene 3D
CDD
ProteinModelPortal
H6VTQ1
C7AQZ9
A0A194R200
A0A194Q1M8
S4PWU7
A0A437B8P7
+ More
A0A212FBN5 A0A2H1W555 A0A2A4JE72 A0A2A4JD33 A0A0L7KL18 A0A0K8TN67 B0W3K6 A0A1Q3FH55 A0A182K4L5 A0A182QQW9 A0A182F942 A0A182PKG2 A0A084W4Y9 A0A2M4BYC0 A0A182T1S0 A0A1D6TIB9 A0A182UPK9 A0A182TSS3 A0A182LAY8 Q5TVD0 A0A182I908 A0A182YP33 A0A182WVC7 W5JB29 A0A067R0M0 T1DM52 A0A2M3ZJN0 A0A182N4K7 A0A182VY17 A0A182M7X9 A0A1I8MRF4 A0A1I8N2P0 A0A1I8MT85 J3JZ42 A0A1I8PFR0 A0A1Y1M290 A0A0T6B8C2 A0A423S9T9 A0A3R7LYD5 Q295I0 B4GDV0 D2A3L8 B4JUS2 A0A336K4I6 A0A2J7PRC0 A0A1A9W8C0 A0A0P4W746 A0A1A9VFM1 A0A0L0C784 A0A1A9XWB1 N6SUV0 A0A1B0FCP7 T1HR47 U4UQX3 A0A1B0A375 B4NJ37 A0A1L8DTW7 A0A1L8DTP6 A0A1L8DU93 A0A0P4VM14 A0A1B6HVP4 A0A1B6FYQ9 E9FYC9 A0A1S3CYK0
A0A212FBN5 A0A2H1W555 A0A2A4JE72 A0A2A4JD33 A0A0L7KL18 A0A0K8TN67 B0W3K6 A0A1Q3FH55 A0A182K4L5 A0A182QQW9 A0A182F942 A0A182PKG2 A0A084W4Y9 A0A2M4BYC0 A0A182T1S0 A0A1D6TIB9 A0A182UPK9 A0A182TSS3 A0A182LAY8 Q5TVD0 A0A182I908 A0A182YP33 A0A182WVC7 W5JB29 A0A067R0M0 T1DM52 A0A2M3ZJN0 A0A182N4K7 A0A182VY17 A0A182M7X9 A0A1I8MRF4 A0A1I8N2P0 A0A1I8MT85 J3JZ42 A0A1I8PFR0 A0A1Y1M290 A0A0T6B8C2 A0A423S9T9 A0A3R7LYD5 Q295I0 B4GDV0 D2A3L8 B4JUS2 A0A336K4I6 A0A2J7PRC0 A0A1A9W8C0 A0A0P4W746 A0A1A9VFM1 A0A0L0C784 A0A1A9XWB1 N6SUV0 A0A1B0FCP7 T1HR47 U4UQX3 A0A1B0A375 B4NJ37 A0A1L8DTW7 A0A1L8DTP6 A0A1L8DU93 A0A0P4VM14 A0A1B6HVP4 A0A1B6FYQ9 E9FYC9 A0A1S3CYK0
PDB
2YUA
E-value=3.45695e-10,
Score=152
Ontologies
KEGG
PANTHER
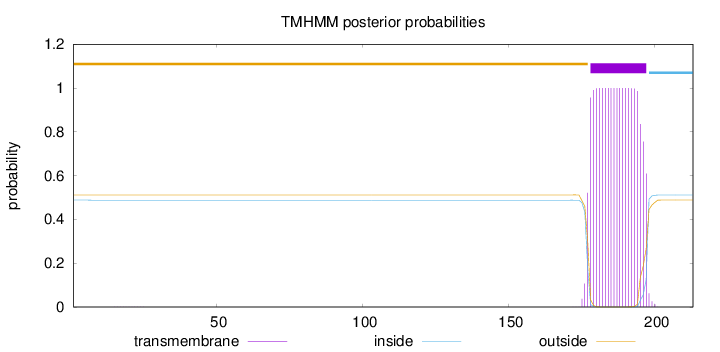
Topology
Length:
213
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.89436
Exp number, first 60 AAs:
0.00253
Total prob of N-in:
0.48813
outside
1 - 177
TMhelix
178 - 197
inside
198 - 213
Population Genetic Test Statistics
Pi
231.266788
Theta
198.891173
Tajima's D
0.474817
CLR
0.05214
CSRT
0.512574371281436
Interpretation
Uncertain
