Gene
KWMTBOMO09668 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012961
Annotation
myosin_light_polypeptide_9_isoform_B_[Bombyx_mori]
Full name
Myosin regulatory light chain sqh
Alternative Name
Myosin regulatory light chain, nonmuscle
Protein spaghetti-squash
Protein spaghetti-squash
Location in the cell
Cytoplasmic Reliability : 1.853 Nuclear Reliability : 1.367
Sequence
CDS
ATGTCATCTCGCAAGACAGCCGGTCGTCGCATTAACAAGAAGCGCGCACAACGTGCAACCTCCAATGTGTTTGCTATGTTTGATCAAGCTCAAATAGCAGAGTTCAAGGAGGCTTTCAATATGATTGATCAGAACAGGGATGGATTCATTGACAAGGACGATCTGCATGATATGCTTGCATCTTTAGGCAAAAACCCAACTGAAGATTATTTAGAGGGAATGATGAATGAAGCACCGGGACCAATTAACTTTACGATGTTTCTCACTCTGTTTGGTGAGAGACTGCAGGGCACTGACCCTGAGGATGTCATCAAAAATGCATTTGGGTGCTTCGATGAAGAGAACAATGGTGTGATTGGTGAGGAGCGACTCCGCGAGCTGCTTACCACAATGGGCGATCGCTTTACAGACGATGATGTGGATGAAATGCTCAGGGAAGCACCAATCCGCGATGGACTCTTCGATTATGTGGAATTCACGCGTATTCTTAAACACGGCGCTAAAGACAAGGACGAGCAATAA
Protein
MSSRKTAGRRINKKRAQRATSNVFAMFDQAQIAEFKEAFNMIDQNRDGFIDKDDLHDMLASLGKNPTEDYLEGMMNEAPGPINFTMFLTLFGERLQGTDPEDVIKNAFGCFDEENNGVIGEERLRELLTTMGDRFTDDDVDEMLREAPIRDGLFDYVEFTRILKHGAKDKDEQ
Summary
Description
Required for cytokinesis, could regulate contractile ring function.
Subunit
Myosin is a hexamer of 2 heavy chains and 4 light chains.
Miscellaneous
This chain binds calcium.
Keywords
Calcium
Complete proteome
Metal-binding
Motor protein
Myosin
Phosphoprotein
Reference proteome
Repeat
Feature
chain Myosin regulatory light chain sqh
Uniprot
Q2F5P3
Q1HPR8
Q2F5P2
I4DQB6
A0A194Q665
A0A2A4JDZ6
+ More
A0A2H1W6X1 A0A194R177 S4P8P3 A0A212FBR6 A0A3S2NGD7 A0A182RKB6 A0A182J7J9 A0A182LT65 A0A182YQ33 A0A182VT34 A0A084VQE2 A0A182YF19 Q7PSW1 A0A182NQE1 A0A182PL39 A0A182TWD2 A0A182LAB6 F5HIZ5 A0A182IDJ3 A0A1Q3G432 A0A1B0CQY0 A0A1Q3G4I4 W8C7Y4 A0A182NVE9 A0A1L8E5K5 A0A182VN16 A0A154NYW8 A0A034WP57 A0A0A1XB80 A0A0K8UTN7 U5EWN4 A0A182X3C7 A0A1A9YJC7 A0A1B0BAX0 A0A1B0A2T8 D3TSF0 A0A182Q1Q2 A0A1Q3G449 A0A0L7QSJ3 Q16XC3 T1E818 A0A2M3Z0Q9 A0A2M4ARN0 A0A2M4C175 W5JRI7 A0A0K8TP86 A0A1A9URI2 T1PBI8 A0A0Q9W781 A0A182QFH9 A0A0M9A6V5 A0A1W4VT32 B4R5D9 A0A3B0JWA2 B4PYP8 B3NXP5 G3GBH2 B4I0E0 Q29GG1 B3MQ57 F6J1D0 B4M865 P40423 F6J1B9 A0A1I8PF28 A0A1L8EH53 A0A1A9W964 Q16XC2 A0A023EGU9 A0A2A3EFZ7 A0A1B6BXB4 K7IN01 A0A088AU38 A0A1W4WK14 B4NPD7 A0A0T6BE92 D6WIC0 A0A2R7W1X3 A0A139WLD1 E9IN39 A0A0A9Y7K4 A0A1B0CZ92 A0A0C9RN65 A0A3L8D8J8 E2ASM0 A0A195BXV0 A0A195BH38 A0A195F1Q4 A0A348G645 E2BVC8 A0A158NL29 A0A026WWH7 A0A151XBM2 F4WFY6
A0A2H1W6X1 A0A194R177 S4P8P3 A0A212FBR6 A0A3S2NGD7 A0A182RKB6 A0A182J7J9 A0A182LT65 A0A182YQ33 A0A182VT34 A0A084VQE2 A0A182YF19 Q7PSW1 A0A182NQE1 A0A182PL39 A0A182TWD2 A0A182LAB6 F5HIZ5 A0A182IDJ3 A0A1Q3G432 A0A1B0CQY0 A0A1Q3G4I4 W8C7Y4 A0A182NVE9 A0A1L8E5K5 A0A182VN16 A0A154NYW8 A0A034WP57 A0A0A1XB80 A0A0K8UTN7 U5EWN4 A0A182X3C7 A0A1A9YJC7 A0A1B0BAX0 A0A1B0A2T8 D3TSF0 A0A182Q1Q2 A0A1Q3G449 A0A0L7QSJ3 Q16XC3 T1E818 A0A2M3Z0Q9 A0A2M4ARN0 A0A2M4C175 W5JRI7 A0A0K8TP86 A0A1A9URI2 T1PBI8 A0A0Q9W781 A0A182QFH9 A0A0M9A6V5 A0A1W4VT32 B4R5D9 A0A3B0JWA2 B4PYP8 B3NXP5 G3GBH2 B4I0E0 Q29GG1 B3MQ57 F6J1D0 B4M865 P40423 F6J1B9 A0A1I8PF28 A0A1L8EH53 A0A1A9W964 Q16XC2 A0A023EGU9 A0A2A3EFZ7 A0A1B6BXB4 K7IN01 A0A088AU38 A0A1W4WK14 B4NPD7 A0A0T6BE92 D6WIC0 A0A2R7W1X3 A0A139WLD1 E9IN39 A0A0A9Y7K4 A0A1B0CZ92 A0A0C9RN65 A0A3L8D8J8 E2ASM0 A0A195BXV0 A0A195BH38 A0A195F1Q4 A0A348G645 E2BVC8 A0A158NL29 A0A026WWH7 A0A151XBM2 F4WFY6
Pubmed
19121390
22651552
26354079
23622113
22118469
25244985
+ More
24438588 12364791 20966253 14747013 17210077 24495485 25348373 25830018 20353571 17510324 20920257 23761445 26369729 25315136 17994087 17550304 21599771 19506309 15632085 23185243 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 21383965 26109357 26109356 1905980 12537569 17372656 18327897 24945155 26483478 20075255 18362917 19820115 21282665 25401762 26823975 30249741 20798317 21347285 24508170 21719571
24438588 12364791 20966253 14747013 17210077 24495485 25348373 25830018 20353571 17510324 20920257 23761445 26369729 25315136 17994087 17550304 21599771 19506309 15632085 23185243 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 21383965 26109357 26109356 1905980 12537569 17372656 18327897 24945155 26483478 20075255 18362917 19820115 21282665 25401762 26823975 30249741 20798317 21347285 24508170 21719571
EMBL
DQ311380
ABD36324.1
DQ443334
ABF51423.1
BABH01003648
BABH01003649
+ More
DQ311381 ABD36325.1 AK404030 BAM20106.1 KQ459582 KPI98900.1 NWSH01001822 PCG70006.1 ODYU01006408 SOQ48224.1 KQ460878 KPJ11548.1 GAIX01009385 JAA83175.1 AGBW02009293 OWR51153.1 RSAL01000115 RVE46881.1 AXCP01007140 AXCM01003211 ATLV01015192 KE525003 KFB40186.1 AAAB01008811 EAA04958.3 EGK96256.1 APCN01004704 GFDL01000473 JAV34572.1 AJWK01024214 GFDL01000366 JAV34679.1 GAMC01000252 JAC06304.1 GFDF01000150 JAV13934.1 KQ434783 KZC04762.1 GAKP01002845 JAC56107.1 GBXI01006237 JAD08055.1 GDHF01033904 GDHF01022614 GDHF01002225 JAI18410.1 JAI29700.1 JAI50089.1 GANO01001339 JAB58532.1 JXJN01011213 CCAG010015760 EZ424352 ADD20628.1 AXCN02001528 GFDL01000481 JAV34564.1 KQ414758 KOC61519.1 CH477543 EAT39249.1 EAT39250.1 GAMD01003133 JAA98457.1 GGFM01001343 MBW22094.1 GGFK01010051 MBW43372.1 GGFJ01009925 MBW59066.1 ADMH02000386 GGFL01004382 ETN66751.1 MBW68560.1 GDAI01001406 JAI16197.1 KA646127 AFP60756.1 CH940653 KRF80562.1 AXCN02002011 KQ435724 KOX78437.1 CM000366 EDX17235.1 OUUW01000011 SPP86334.1 CM000162 EDX00984.1 CH954180 EDV47346.1 JF815874 JF815875 JF815876 JF815877 JF815878 JF815879 JF815880 JF815881 JF815882 JF815883 JF815884 JF815885 JF815886 JF815887 JF815888 JF815889 JF815890 JF815891 JF815892 JF815893 JF815894 JF815895 JF815896 JF815897 JF815898 JF815899 JF815900 JF815901 JF815902 JF815903 JF815904 JF815905 JF815906 JF815907 JF815908 JF815909 JF815910 JF815911 JF815912 JF815913 JF815914 JF815915 AEN71309.1 FJ696192 FJ696193 FJ696194 FJ696195 FJ696196 FJ696197 FJ696198 FJ696199 FJ696200 FJ696201 FJ696202 CH480819 ACV81714.1 EDW52971.1 CH379064 EAL32148.1 KRT06998.1 CH902621 EDV44483.1 KPU81615.1 GQ306651 GQ306652 GQ306653 GQ306654 GQ306655 GQ306656 GQ306657 GQ306658 GQ306659 GQ306660 GQ306661 GQ306662 GQ306663 GQ306664 GQ306665 GQ306666 GQ306667 GQ306668 GQ306669 GQ306670 GQ306671 GQ306672 GQ306673 AE014298 ADG51498.1 ADG51499.1 ADG51500.1 ADG51501.1 ADG51502.1 ADG51503.1 ADG51504.1 ADG51505.1 ADG51506.1 ADG51507.1 ADG51508.1 ADG51509.1 ADG51510.1 ADG51511.1 ADG51512.1 ADG51513.1 ADG51514.1 ADG51515.1 ADG51516.1 ADG51517.1 ADG51518.1 ADG51519.1 ADG51520.1 AHN59399.1 AHN59400.1 AHN59401.1 AHN59402.1 CH941246 EDW62341.1 EDW71474.1 M67494 AY122159 GQ306650 ADG51497.1 GFDG01000747 JAV18052.1 EAT39251.1 EAT39252.1 JXUM01022978 JXUM01022979 JXUM01022980 JXUM01022981 JXUM01079721 JXUM01079722 JXUM01079723 JXUM01079724 GAPW01005034 GAPW01005033 GAPW01005031 GAPW01005030 JAC08568.1 KZ288255 PBC30675.1 GEDC01031371 JAS05927.1 CH964291 EDW86377.1 LJIG01001371 KRT85569.1 KQ971321 EFA01437.1 KK854252 PTY13747.1 KYB28742.1 GL764272 EFZ18005.1 GBHO01015470 GDHC01016192 JAG28134.1 JAQ02437.1 AJVK01002143 AJVK01002144 AJVK01002145 AJVK01002146 GBYB01002520 GBYB01014922 JAG72287.1 JAG84689.1 QOIP01000011 RLU16413.1 GL442330 EFN63580.1 KQ978501 KYM93484.1 KQ976488 KYM83543.1 KQ981866 KYN34102.1 FX985585 BBF97918.1 GL450824 EFN80420.1 ADTU01019229 KK107078 EZA60407.1 KQ982320 KYQ57754.1 GL888128 EGI66753.1
DQ311381 ABD36325.1 AK404030 BAM20106.1 KQ459582 KPI98900.1 NWSH01001822 PCG70006.1 ODYU01006408 SOQ48224.1 KQ460878 KPJ11548.1 GAIX01009385 JAA83175.1 AGBW02009293 OWR51153.1 RSAL01000115 RVE46881.1 AXCP01007140 AXCM01003211 ATLV01015192 KE525003 KFB40186.1 AAAB01008811 EAA04958.3 EGK96256.1 APCN01004704 GFDL01000473 JAV34572.1 AJWK01024214 GFDL01000366 JAV34679.1 GAMC01000252 JAC06304.1 GFDF01000150 JAV13934.1 KQ434783 KZC04762.1 GAKP01002845 JAC56107.1 GBXI01006237 JAD08055.1 GDHF01033904 GDHF01022614 GDHF01002225 JAI18410.1 JAI29700.1 JAI50089.1 GANO01001339 JAB58532.1 JXJN01011213 CCAG010015760 EZ424352 ADD20628.1 AXCN02001528 GFDL01000481 JAV34564.1 KQ414758 KOC61519.1 CH477543 EAT39249.1 EAT39250.1 GAMD01003133 JAA98457.1 GGFM01001343 MBW22094.1 GGFK01010051 MBW43372.1 GGFJ01009925 MBW59066.1 ADMH02000386 GGFL01004382 ETN66751.1 MBW68560.1 GDAI01001406 JAI16197.1 KA646127 AFP60756.1 CH940653 KRF80562.1 AXCN02002011 KQ435724 KOX78437.1 CM000366 EDX17235.1 OUUW01000011 SPP86334.1 CM000162 EDX00984.1 CH954180 EDV47346.1 JF815874 JF815875 JF815876 JF815877 JF815878 JF815879 JF815880 JF815881 JF815882 JF815883 JF815884 JF815885 JF815886 JF815887 JF815888 JF815889 JF815890 JF815891 JF815892 JF815893 JF815894 JF815895 JF815896 JF815897 JF815898 JF815899 JF815900 JF815901 JF815902 JF815903 JF815904 JF815905 JF815906 JF815907 JF815908 JF815909 JF815910 JF815911 JF815912 JF815913 JF815914 JF815915 AEN71309.1 FJ696192 FJ696193 FJ696194 FJ696195 FJ696196 FJ696197 FJ696198 FJ696199 FJ696200 FJ696201 FJ696202 CH480819 ACV81714.1 EDW52971.1 CH379064 EAL32148.1 KRT06998.1 CH902621 EDV44483.1 KPU81615.1 GQ306651 GQ306652 GQ306653 GQ306654 GQ306655 GQ306656 GQ306657 GQ306658 GQ306659 GQ306660 GQ306661 GQ306662 GQ306663 GQ306664 GQ306665 GQ306666 GQ306667 GQ306668 GQ306669 GQ306670 GQ306671 GQ306672 GQ306673 AE014298 ADG51498.1 ADG51499.1 ADG51500.1 ADG51501.1 ADG51502.1 ADG51503.1 ADG51504.1 ADG51505.1 ADG51506.1 ADG51507.1 ADG51508.1 ADG51509.1 ADG51510.1 ADG51511.1 ADG51512.1 ADG51513.1 ADG51514.1 ADG51515.1 ADG51516.1 ADG51517.1 ADG51518.1 ADG51519.1 ADG51520.1 AHN59399.1 AHN59400.1 AHN59401.1 AHN59402.1 CH941246 EDW62341.1 EDW71474.1 M67494 AY122159 GQ306650 ADG51497.1 GFDG01000747 JAV18052.1 EAT39251.1 EAT39252.1 JXUM01022978 JXUM01022979 JXUM01022980 JXUM01022981 JXUM01079721 JXUM01079722 JXUM01079723 JXUM01079724 GAPW01005034 GAPW01005033 GAPW01005031 GAPW01005030 JAC08568.1 KZ288255 PBC30675.1 GEDC01031371 JAS05927.1 CH964291 EDW86377.1 LJIG01001371 KRT85569.1 KQ971321 EFA01437.1 KK854252 PTY13747.1 KYB28742.1 GL764272 EFZ18005.1 GBHO01015470 GDHC01016192 JAG28134.1 JAQ02437.1 AJVK01002143 AJVK01002144 AJVK01002145 AJVK01002146 GBYB01002520 GBYB01014922 JAG72287.1 JAG84689.1 QOIP01000011 RLU16413.1 GL442330 EFN63580.1 KQ978501 KYM93484.1 KQ976488 KYM83543.1 KQ981866 KYN34102.1 FX985585 BBF97918.1 GL450824 EFN80420.1 ADTU01019229 KK107078 EZA60407.1 KQ982320 KYQ57754.1 GL888128 EGI66753.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000007151
UP000283053
+ More
UP000075900 UP000075880 UP000075883 UP000076408 UP000075920 UP000030765 UP000007062 UP000075884 UP000075885 UP000075902 UP000075882 UP000075840 UP000092461 UP000075903 UP000076502 UP000076407 UP000092443 UP000092460 UP000092445 UP000092444 UP000075886 UP000053825 UP000008820 UP000000673 UP000078200 UP000095301 UP000008792 UP000053105 UP000192221 UP000000304 UP000268350 UP000002282 UP000008711 UP000001292 UP000001819 UP000007801 UP000000803 UP000095300 UP000091820 UP000069940 UP000242457 UP000002358 UP000005203 UP000192223 UP000007798 UP000007266 UP000092462 UP000279307 UP000000311 UP000078542 UP000078540 UP000078541 UP000008237 UP000005205 UP000053097 UP000075809 UP000007755
UP000075900 UP000075880 UP000075883 UP000076408 UP000075920 UP000030765 UP000007062 UP000075884 UP000075885 UP000075902 UP000075882 UP000075840 UP000092461 UP000075903 UP000076502 UP000076407 UP000092443 UP000092460 UP000092445 UP000092444 UP000075886 UP000053825 UP000008820 UP000000673 UP000078200 UP000095301 UP000008792 UP000053105 UP000192221 UP000000304 UP000268350 UP000002282 UP000008711 UP000001292 UP000001819 UP000007801 UP000000803 UP000095300 UP000091820 UP000069940 UP000242457 UP000002358 UP000005203 UP000192223 UP000007798 UP000007266 UP000092462 UP000279307 UP000000311 UP000078542 UP000078540 UP000078541 UP000008237 UP000005205 UP000053097 UP000075809 UP000007755
Pfam
Interpro
IPR018247
EF_Hand_1_Ca_BS
+ More
IPR011992 EF-hand-dom_pair
IPR002048 EF_hand_dom
IPR035976 Sushi/SCR/CCP_sf
IPR001304 C-type_lectin-like
IPR006585 FTP1
IPR016186 C-type_lectin-like/link_sf
IPR000421 FA58C
IPR008979 Galactose-bd-like_sf
IPR016187 CTDL_fold
IPR000436 Sushi_SCR_CCP_dom
IPR018378 C-type_lectin_CS
IPR011992 EF-hand-dom_pair
IPR002048 EF_hand_dom
IPR035976 Sushi/SCR/CCP_sf
IPR001304 C-type_lectin-like
IPR006585 FTP1
IPR016186 C-type_lectin-like/link_sf
IPR000421 FA58C
IPR008979 Galactose-bd-like_sf
IPR016187 CTDL_fold
IPR000436 Sushi_SCR_CCP_dom
IPR018378 C-type_lectin_CS
Gene 3D
ProteinModelPortal
Q2F5P3
Q1HPR8
Q2F5P2
I4DQB6
A0A194Q665
A0A2A4JDZ6
+ More
A0A2H1W6X1 A0A194R177 S4P8P3 A0A212FBR6 A0A3S2NGD7 A0A182RKB6 A0A182J7J9 A0A182LT65 A0A182YQ33 A0A182VT34 A0A084VQE2 A0A182YF19 Q7PSW1 A0A182NQE1 A0A182PL39 A0A182TWD2 A0A182LAB6 F5HIZ5 A0A182IDJ3 A0A1Q3G432 A0A1B0CQY0 A0A1Q3G4I4 W8C7Y4 A0A182NVE9 A0A1L8E5K5 A0A182VN16 A0A154NYW8 A0A034WP57 A0A0A1XB80 A0A0K8UTN7 U5EWN4 A0A182X3C7 A0A1A9YJC7 A0A1B0BAX0 A0A1B0A2T8 D3TSF0 A0A182Q1Q2 A0A1Q3G449 A0A0L7QSJ3 Q16XC3 T1E818 A0A2M3Z0Q9 A0A2M4ARN0 A0A2M4C175 W5JRI7 A0A0K8TP86 A0A1A9URI2 T1PBI8 A0A0Q9W781 A0A182QFH9 A0A0M9A6V5 A0A1W4VT32 B4R5D9 A0A3B0JWA2 B4PYP8 B3NXP5 G3GBH2 B4I0E0 Q29GG1 B3MQ57 F6J1D0 B4M865 P40423 F6J1B9 A0A1I8PF28 A0A1L8EH53 A0A1A9W964 Q16XC2 A0A023EGU9 A0A2A3EFZ7 A0A1B6BXB4 K7IN01 A0A088AU38 A0A1W4WK14 B4NPD7 A0A0T6BE92 D6WIC0 A0A2R7W1X3 A0A139WLD1 E9IN39 A0A0A9Y7K4 A0A1B0CZ92 A0A0C9RN65 A0A3L8D8J8 E2ASM0 A0A195BXV0 A0A195BH38 A0A195F1Q4 A0A348G645 E2BVC8 A0A158NL29 A0A026WWH7 A0A151XBM2 F4WFY6
A0A2H1W6X1 A0A194R177 S4P8P3 A0A212FBR6 A0A3S2NGD7 A0A182RKB6 A0A182J7J9 A0A182LT65 A0A182YQ33 A0A182VT34 A0A084VQE2 A0A182YF19 Q7PSW1 A0A182NQE1 A0A182PL39 A0A182TWD2 A0A182LAB6 F5HIZ5 A0A182IDJ3 A0A1Q3G432 A0A1B0CQY0 A0A1Q3G4I4 W8C7Y4 A0A182NVE9 A0A1L8E5K5 A0A182VN16 A0A154NYW8 A0A034WP57 A0A0A1XB80 A0A0K8UTN7 U5EWN4 A0A182X3C7 A0A1A9YJC7 A0A1B0BAX0 A0A1B0A2T8 D3TSF0 A0A182Q1Q2 A0A1Q3G449 A0A0L7QSJ3 Q16XC3 T1E818 A0A2M3Z0Q9 A0A2M4ARN0 A0A2M4C175 W5JRI7 A0A0K8TP86 A0A1A9URI2 T1PBI8 A0A0Q9W781 A0A182QFH9 A0A0M9A6V5 A0A1W4VT32 B4R5D9 A0A3B0JWA2 B4PYP8 B3NXP5 G3GBH2 B4I0E0 Q29GG1 B3MQ57 F6J1D0 B4M865 P40423 F6J1B9 A0A1I8PF28 A0A1L8EH53 A0A1A9W964 Q16XC2 A0A023EGU9 A0A2A3EFZ7 A0A1B6BXB4 K7IN01 A0A088AU38 A0A1W4WK14 B4NPD7 A0A0T6BE92 D6WIC0 A0A2R7W1X3 A0A139WLD1 E9IN39 A0A0A9Y7K4 A0A1B0CZ92 A0A0C9RN65 A0A3L8D8J8 E2ASM0 A0A195BXV0 A0A195BH38 A0A195F1Q4 A0A348G645 E2BVC8 A0A158NL29 A0A026WWH7 A0A151XBM2 F4WFY6
PDB
3J04
E-value=1.51447e-57,
Score=559
Ontologies
GO
GO:0005509
GO:0110069
GO:0000281
GO:0035317
GO:0035159
GO:0005912
GO:0048471
GO:0042060
GO:0035148
GO:0019749
GO:1903144
GO:0005911
GO:0060288
GO:0003384
GO:0035191
GO:0060289
GO:0035277
GO:0035183
GO:0016324
GO:0090254
GO:0016460
GO:0031036
GO:0007298
GO:0110071
GO:0032036
GO:0106037
GO:0030496
GO:0051233
GO:0005829
GO:0032956
GO:0045177
GO:0005737
GO:0032154
GO:0001736
GO:0045179
GO:0005938
GO:0007300
GO:0005886
GO:0030707
GO:0030048
GO:0016021
GO:0050660
GO:0016491
GO:0006508
GO:0016787
GO:0003723
GO:0006730
GO:0009258
GO:0016155
GO:0003824
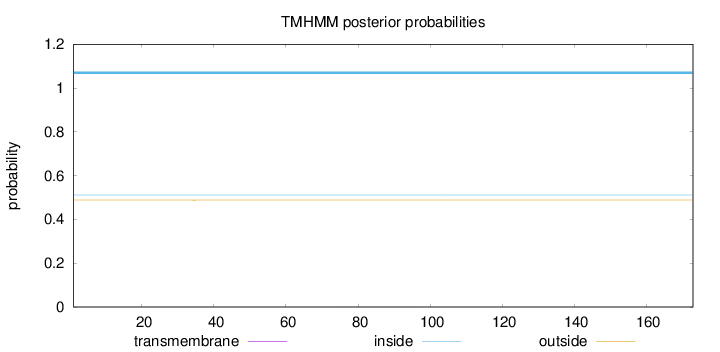
Topology
Length:
173
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01146
Exp number, first 60 AAs:
0.00636
Total prob of N-in:
0.51178
inside
1 - 173
Population Genetic Test Statistics
Pi
187.596808
Theta
185.654566
Tajima's D
0.565559
CLR
28.989358
CSRT
0.530623468826559
Interpretation
Uncertain
