Gene
KWMTBOMO09667 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012872
Annotation
PREDICTED:_glucose_dehydrogenase_[FAD?_quinone]-like_[Amyelois_transitella]
Full name
Endoglucanase
+ More
Glucose dehydrogenase [FAD, quinone]
Glucose dehydrogenase [FAD, quinone]
Location in the cell
Cytoplasmic Reliability : 1.97
Sequence
CDS
ATGATATTAGCTCCAGAGGAGTGCGGCTGTCCGCTTATTGAAGAAGGAGTCTCTATAGAAAACTCGCCGATTTGCAGTGGAACTCTGCTATTTATGGTATTACTCGAAGGGTACATCCGAGGTCGTTGCAAAATCGCAACTCCATGCGCTAATGTGAGATCCGCTGAACAACTGGATGCCCGTTACGATTTTATTGTGGTGGGCTCAGGTCCGGCCGGAGCCATTGTTGCTGGAAGACTCAGCGAAGATAAGAATTTTAATGTACTATTACTGGAAGCCGGAGGTCAAGAGCCTACGGGAGCACGCATTCCCTCTTTCTACAGAGCATTCTGGTCGAACAAAGAGGTGGATTGGGATTATAGAACTGAGCCCGACAACTATTGTCTCGACCAAGGCGATAAAGGATGTCTTTGGCCACGAGGAAAGGTTCTTGGTGGATCAAGTCTCCTTAATGGTATGATGTACCATCGAGGCCATGCCGCTGATTACGATGACTGGGTGAAGCTCGGTGCTGAAGGTTGGTCATGGGAGGAAAATCTTCCATACTTTGACATGACCGAGGGGAACAAGGAAATTGGAACTATAGTGAGCGAGAAACACCACTCTAGTTCAGGGCCTTTGCCTGTTCAAAGATTCCGTTACCAAGGTCCAGCCGTTTATAAACTATTGGATGCCCTCAACGAAACTGGGTTCTCTATCATAGCTGACATGAACGATCCTGAAACTCCCGATGGATTCACCATAGCACAAGCGTTTAACGATAACGGCCAGAGGTACACAACAGCGCGCGCGTACCTGAAACCGAAGTCAGAACGACCGAACCTGACAGTGAAACTGGGAGCGCACGTTACAAGAGTCATAGTGACTGACGACACAGCCACCGGCGTTGAGTTCATCGACTCCGACGGAAATTCAAATATCGTTTATGCTTCTCGAGAGGTGATTTTAAGTGCTGGAGCGTTAAACACTCCTCACATTCTTTTGCATTCCGGCATTGGACCCCGAGAGACGCTAGAGAAGTACAACATTCCAGTCAAAGCAGATCTCCCAGTGGGTTTGAATCTTCAGAATCACGTTGGTGTCACTGTATCGTTTATACTGCCTAAACTCAACGACACACGAGTCTTGGATTGGAGCACGCTTGCCACATACTTGTTGAATCAGGAGGGGCCTATGACGTCCACCTCAATTACTCAGGTAACAGGATTGTTGTATTCAAGTTTGGCGGACAAAAGAAAAAAGCAACCTGACCTTCAATTCTTCTTTAATGGCATGTATGCAGAATGCTCCAAGACGGGCTTCGTGGGAGAGACTATTGACAGTGACTGCCAAGAAAGAGGAACAAATATTACAGCCAATGCAGTGGCGCTCCTGCCAAAAAGCAAGGGATACTTAACGTTGCAGTCTTCAGACCCACTAGTTTCTCCATTATTTTATCCGAATTTCTTCTCACATCCTGATGATATGATCGTGGCCAAAGACGGACTCAGATATTTGAAAAAAATATCTGAAAGCAAGATCCTAGAATCAGAATATGGCATAGAACTAGATCCCGAAGCGACTGATGAGTGTAGTGAGACCTCAGAAGACTGGTCAGATGACTGGATGGAGTGCATGATCCGATTGCACACTGATGCCCAGAATCATCAACTCGGCACAACCGCTATCGGCTTGGTCGTCGATCCTCAACTAAAAGTCTACGGAATTGAAGGTTTGAGAGTTATCGATGCATCGGTCATGCCGTCACAGCCGACGGGCAATCCTCAAGCAGCGATCATGATGGTCGCCGAGCGAGGCGCGGCTTTCATCAAAGACACTCATTCCTAA
Protein
MILAPEECGCPLIEEGVSIENSPICSGTLLFMVLLEGYIRGRCKIATPCANVRSAEQLDARYDFIVVGSGPAGAIVAGRLSEDKNFNVLLLEAGGQEPTGARIPSFYRAFWSNKEVDWDYRTEPDNYCLDQGDKGCLWPRGKVLGGSSLLNGMMYHRGHAADYDDWVKLGAEGWSWEENLPYFDMTEGNKEIGTIVSEKHHSSSGPLPVQRFRYQGPAVYKLLDALNETGFSIIADMNDPETPDGFTIAQAFNDNGQRYTTARAYLKPKSERPNLTVKLGAHVTRVIVTDDTATGVEFIDSDGNSNIVYASREVILSAGALNTPHILLHSGIGPRETLEKYNIPVKADLPVGLNLQNHVGVTVSFILPKLNDTRVLDWSTLATYLLNQEGPMTSTSITQVTGLLYSSLADKRKKQPDLQFFFNGMYAECSKTGFVGETIDSDCQERGTNITANAVALLPKSKGYLTLQSSDPLVSPLFYPNFFSHPDDMIVAKDGLRYLKKISESKILESEYGIELDPEATDECSETSEDWSDDWMECMIRLHTDAQNHQLGTTAIGLVVDPQLKVYGIEGLRVIDASVMPSQPTGNPQAAIMMVAERGAAFIKDTHS
Summary
Catalytic Activity
Endohydrolysis of (1->4)-beta-D-glucosidic linkages in cellulose, lichenin and cereal beta-D-glucans.
a quinone + D-glucose = a quinol + D-glucono-1,5-lactone
a quinone + D-glucose = a quinol + D-glucono-1,5-lactone
Cofactor
FAD
Similarity
Belongs to the GMC oxidoreductase family.
Belongs to the glycosyl hydrolase 9 (cellulase E) family.
Belongs to the glycosyl hydrolase 9 (cellulase E) family.
Keywords
Complete proteome
FAD
Flavoprotein
Oxidoreductase
Reference proteome
Secreted
Selenocysteine
Signal
Feature
chain Endoglucanase
Uniprot
H9JTL0
A0A437B8Q7
A0A142I707
D9ZFI1
A0A2H1W5D5
B2MW81
+ More
B8XZY7 A0A194Q009 A0A2A4JD42 A0A218N0E8 A0A212FMI8 A0A2H1VAE8 A0A194RR25 A0A194Q0T0 A0A3S2LZ99 A0A2A4K284 A0A2S2NJV8 A0A182RS69 A0A2M4CT25 J9JWH7 D6WPB8 A0A182VBZ4 A0A182XAV1 A0A182LES2 Q7QCC5 A0A182HSP9 A0A182MWU5 A0A1Y1KHY8 A0A182VWW9 A0A182UL69 E0VK19 A0A139WGK8 A0A1Q3FML5 A0A182J274 A0A087ZQ05 B0WR76 A0A182NRL5 A0A2H1WCL9 A0A2A3EJY8 A0A182R0I2 A0A2J7Q741 A0A232F9V9 E2AQ44 A0A3L8E234 A0A067R3T3 A0A291P102 A0A336MPQ2 A0A1B6EWY6 A0A336ME48 A0A1I8PT77 K7ITZ6 A0A2J7QLL4 A0A291P0R7 A0A158NWQ4 A0A2P8XMK6 A0A212F246 A0A1S3D8R6 Q178U1 A0A2S2QBR1 A0A437BLQ3 F4WQ30 A0A0A9ZH27 A0A151IHR1 A0A1A9VQK6 A0A023EV68 A0A194PXW0 A0A067QZE3 B4GMR4 D0QWK8 A0A1I8N555 A0A1A9Y074 A0A0R3NJP9 A0A1B0FAV7 A0A1A9Z6K6 A0A1Y1MP57 P18172 A0A026W0A3 A0A3B0JPT6 D6WN48 O44106 A0A151JUB7 A0A154P559 B4M4G1 A0A195EDG1 A0A1A9WL86 A0A1B0DAQ7 B3LYF6 Q8SXV0 V5GRM6 A0A151I5L3 B4I4R7 E9IVZ8 A0A0C9S068 U3PT72 Q94963 A0A2U8U0K3 P18173 B3P2V7 A0A0C9PXZ5 B4N874
B8XZY7 A0A194Q009 A0A2A4JD42 A0A218N0E8 A0A212FMI8 A0A2H1VAE8 A0A194RR25 A0A194Q0T0 A0A3S2LZ99 A0A2A4K284 A0A2S2NJV8 A0A182RS69 A0A2M4CT25 J9JWH7 D6WPB8 A0A182VBZ4 A0A182XAV1 A0A182LES2 Q7QCC5 A0A182HSP9 A0A182MWU5 A0A1Y1KHY8 A0A182VWW9 A0A182UL69 E0VK19 A0A139WGK8 A0A1Q3FML5 A0A182J274 A0A087ZQ05 B0WR76 A0A182NRL5 A0A2H1WCL9 A0A2A3EJY8 A0A182R0I2 A0A2J7Q741 A0A232F9V9 E2AQ44 A0A3L8E234 A0A067R3T3 A0A291P102 A0A336MPQ2 A0A1B6EWY6 A0A336ME48 A0A1I8PT77 K7ITZ6 A0A2J7QLL4 A0A291P0R7 A0A158NWQ4 A0A2P8XMK6 A0A212F246 A0A1S3D8R6 Q178U1 A0A2S2QBR1 A0A437BLQ3 F4WQ30 A0A0A9ZH27 A0A151IHR1 A0A1A9VQK6 A0A023EV68 A0A194PXW0 A0A067QZE3 B4GMR4 D0QWK8 A0A1I8N555 A0A1A9Y074 A0A0R3NJP9 A0A1B0FAV7 A0A1A9Z6K6 A0A1Y1MP57 P18172 A0A026W0A3 A0A3B0JPT6 D6WN48 O44106 A0A151JUB7 A0A154P559 B4M4G1 A0A195EDG1 A0A1A9WL86 A0A1B0DAQ7 B3LYF6 Q8SXV0 V5GRM6 A0A151I5L3 B4I4R7 E9IVZ8 A0A0C9S068 U3PT72 Q94963 A0A2U8U0K3 P18173 B3P2V7 A0A0C9PXZ5 B4N874
EC Number
3.2.1.4
1.1.5.9
1.1.5.9
Pubmed
19121390
20688075
26354079
22118469
18362917
19820115
+ More
20966253 12364791 14747013 17210077 28004739 20566863 28648823 20798317 30249741 24845553 28986331 20075255 21347285 29403074 17510324 21719571 25401762 26823975 24945155 17994087 19859648 25315136 15632085 23185243 2108306 15545653 24508170 9720289 21282665 8865667 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 3143620 2002763
20966253 12364791 14747013 17210077 28004739 20566863 28648823 20798317 30249741 24845553 28986331 20075255 21347285 29403074 17510324 21719571 25401762 26823975 24945155 17994087 19859648 25315136 15632085 23185243 2108306 15545653 24508170 9720289 21282665 8865667 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 3143620 2002763
EMBL
BABH01003649
BABH01003650
RSAL01000115
RVE46880.1
KT907054
AMR44226.1
+ More
GU983912 ADL38963.1 ODYU01006408 SOQ48223.1 EU629216 ACC94296.1 FJ460711 ACJ71598.1 KQ459582 KPI98901.1 NWSH01001822 PCG70005.1 KY348779 ASF79657.1 AGBW02007651 OWR54951.1 ODYU01001490 SOQ37766.1 KQ459700 KPJ20333.1 KPI98918.1 RSAL01000102 RVE47449.1 NWSH01000252 PCG78014.1 GGMR01004840 MBY17459.1 GGFL01004328 MBW68506.1 ABLF02025721 KQ971354 EFA07378.1 AAAB01008859 EAA08043.3 APCN01000318 AXCM01004736 GEZM01085307 JAV60011.1 DS235237 EEB13725.1 KQ971344 KYB27092.1 GFDL01006281 JAV28764.1 DS232052 EDS33250.1 ODYU01007445 SOQ50224.1 KZ288227 PBC31794.1 AXCN02000004 NEVH01017447 PNF24406.1 NNAY01000667 OXU27097.1 GL441691 EFN64444.1 QOIP01000001 RLU26592.1 KK852777 KDR16732.1 MF687604 ATJ44530.1 UFQS01001751 UFQT01001751 SSX12203.1 SSX31655.1 GECZ01027399 JAS42370.1 UFQT01000848 SSX27631.1 NEVH01013243 PNF29476.1 MF687548 ATJ44474.1 ADTU01002979 PYGN01001713 PSN33241.1 AGBW02010783 OWR47815.1 CH477359 EAT42711.1 GGMS01005964 MBY75167.1 RSAL01000036 RVE51239.1 GL888262 EGI63653.1 GBHO01000088 GBHO01000087 GBRD01012593 GDHC01011845 GDHC01008977 JAG43516.1 JAG43517.1 JAG53231.1 JAQ06784.1 JAQ09652.1 KQ977586 KYN01695.1 GAPW01000590 JAC13008.1 KQ459595 KPI95985.1 KK853157 KDR10473.1 CH479185 EDW38138.1 FJ821032 ACN94707.1 CM000070 KRT01170.1 KRT01171.1 CCAG010022189 GEZM01025416 JAV87459.1 M29299 AY754494 KK107522 EZA49345.1 OUUW01000008 SPP84125.1 KQ971342 EFA03070.1 AF025811 AAB87896.1 KQ981799 KYN35699.1 KQ434809 KZC06464.1 CH940652 EDW59522.2 KQ979074 KYN22852.1 AJVK01029227 CH902617 EDV41819.2 AY084122 AAL89860.1 GALX01004184 JAB64282.1 KQ976434 KYM87196.1 CH480821 EDW55210.1 GL766449 EFZ15289.1 GBYB01013861 JAG83628.1 BT150358 AGW52167.1 AH005098 U63324 AAB48020.1 AE014297 AWM95310.1 M29298 X13582 CH954181 EDV48129.1 GBYB01006377 GBYB01006378 GBYB01006379 GBYB01006380 GBYB01011403 JAG76144.1 JAG76145.1 JAG76146.1 JAG76147.1 JAG81170.1 CH964232 EDW81325.2
GU983912 ADL38963.1 ODYU01006408 SOQ48223.1 EU629216 ACC94296.1 FJ460711 ACJ71598.1 KQ459582 KPI98901.1 NWSH01001822 PCG70005.1 KY348779 ASF79657.1 AGBW02007651 OWR54951.1 ODYU01001490 SOQ37766.1 KQ459700 KPJ20333.1 KPI98918.1 RSAL01000102 RVE47449.1 NWSH01000252 PCG78014.1 GGMR01004840 MBY17459.1 GGFL01004328 MBW68506.1 ABLF02025721 KQ971354 EFA07378.1 AAAB01008859 EAA08043.3 APCN01000318 AXCM01004736 GEZM01085307 JAV60011.1 DS235237 EEB13725.1 KQ971344 KYB27092.1 GFDL01006281 JAV28764.1 DS232052 EDS33250.1 ODYU01007445 SOQ50224.1 KZ288227 PBC31794.1 AXCN02000004 NEVH01017447 PNF24406.1 NNAY01000667 OXU27097.1 GL441691 EFN64444.1 QOIP01000001 RLU26592.1 KK852777 KDR16732.1 MF687604 ATJ44530.1 UFQS01001751 UFQT01001751 SSX12203.1 SSX31655.1 GECZ01027399 JAS42370.1 UFQT01000848 SSX27631.1 NEVH01013243 PNF29476.1 MF687548 ATJ44474.1 ADTU01002979 PYGN01001713 PSN33241.1 AGBW02010783 OWR47815.1 CH477359 EAT42711.1 GGMS01005964 MBY75167.1 RSAL01000036 RVE51239.1 GL888262 EGI63653.1 GBHO01000088 GBHO01000087 GBRD01012593 GDHC01011845 GDHC01008977 JAG43516.1 JAG43517.1 JAG53231.1 JAQ06784.1 JAQ09652.1 KQ977586 KYN01695.1 GAPW01000590 JAC13008.1 KQ459595 KPI95985.1 KK853157 KDR10473.1 CH479185 EDW38138.1 FJ821032 ACN94707.1 CM000070 KRT01170.1 KRT01171.1 CCAG010022189 GEZM01025416 JAV87459.1 M29299 AY754494 KK107522 EZA49345.1 OUUW01000008 SPP84125.1 KQ971342 EFA03070.1 AF025811 AAB87896.1 KQ981799 KYN35699.1 KQ434809 KZC06464.1 CH940652 EDW59522.2 KQ979074 KYN22852.1 AJVK01029227 CH902617 EDV41819.2 AY084122 AAL89860.1 GALX01004184 JAB64282.1 KQ976434 KYM87196.1 CH480821 EDW55210.1 GL766449 EFZ15289.1 GBYB01013861 JAG83628.1 BT150358 AGW52167.1 AH005098 U63324 AAB48020.1 AE014297 AWM95310.1 M29298 X13582 CH954181 EDV48129.1 GBYB01006377 GBYB01006378 GBYB01006379 GBYB01006380 GBYB01011403 JAG76144.1 JAG76145.1 JAG76146.1 JAG76147.1 JAG81170.1 CH964232 EDW81325.2
Proteomes
UP000005204
UP000283053
UP000053268
UP000218220
UP000007151
UP000053240
+ More
UP000075900 UP000007819 UP000007266 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000075883 UP000075920 UP000075902 UP000009046 UP000075880 UP000005203 UP000002320 UP000075884 UP000242457 UP000075886 UP000235965 UP000215335 UP000000311 UP000279307 UP000027135 UP000095300 UP000002358 UP000005205 UP000245037 UP000079169 UP000008820 UP000007755 UP000078542 UP000078200 UP000008744 UP000095301 UP000092443 UP000001819 UP000092444 UP000092445 UP000053097 UP000268350 UP000078541 UP000076502 UP000008792 UP000078492 UP000091820 UP000092462 UP000007801 UP000078540 UP000001292 UP000000803 UP000008711 UP000007798
UP000075900 UP000007819 UP000007266 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000075883 UP000075920 UP000075902 UP000009046 UP000075880 UP000005203 UP000002320 UP000075884 UP000242457 UP000075886 UP000235965 UP000215335 UP000000311 UP000279307 UP000027135 UP000095300 UP000002358 UP000005205 UP000245037 UP000079169 UP000008820 UP000007755 UP000078542 UP000078200 UP000008744 UP000095301 UP000092443 UP000001819 UP000092444 UP000092445 UP000053097 UP000268350 UP000078541 UP000076502 UP000008792 UP000078492 UP000091820 UP000092462 UP000007801 UP000078540 UP000001292 UP000000803 UP000008711 UP000007798
Interpro
IPR036188
FAD/NAD-bd_sf
+ More
IPR000172 GMC_OxRdtase_N
IPR012132 GMC_OxRdtase
IPR007867 GMC_OxRtase_C
IPR027901 DUF4541
IPR027424 Glucose_Oxidase_domain_2
IPR008928 6-hairpin_glycosidase_sf
IPR018221 Glyco_hydro_9_His_AS
IPR012341 6hp_glycosidase-like_sf
IPR033126 Glyco_hydro_9_Asp/Glu_AS
IPR001701 Glyco_hydro_9
IPR000172 GMC_OxRdtase_N
IPR012132 GMC_OxRdtase
IPR007867 GMC_OxRtase_C
IPR027901 DUF4541
IPR027424 Glucose_Oxidase_domain_2
IPR008928 6-hairpin_glycosidase_sf
IPR018221 Glyco_hydro_9_His_AS
IPR012341 6hp_glycosidase-like_sf
IPR033126 Glyco_hydro_9_Asp/Glu_AS
IPR001701 Glyco_hydro_9
Gene 3D
ProteinModelPortal
H9JTL0
A0A437B8Q7
A0A142I707
D9ZFI1
A0A2H1W5D5
B2MW81
+ More
B8XZY7 A0A194Q009 A0A2A4JD42 A0A218N0E8 A0A212FMI8 A0A2H1VAE8 A0A194RR25 A0A194Q0T0 A0A3S2LZ99 A0A2A4K284 A0A2S2NJV8 A0A182RS69 A0A2M4CT25 J9JWH7 D6WPB8 A0A182VBZ4 A0A182XAV1 A0A182LES2 Q7QCC5 A0A182HSP9 A0A182MWU5 A0A1Y1KHY8 A0A182VWW9 A0A182UL69 E0VK19 A0A139WGK8 A0A1Q3FML5 A0A182J274 A0A087ZQ05 B0WR76 A0A182NRL5 A0A2H1WCL9 A0A2A3EJY8 A0A182R0I2 A0A2J7Q741 A0A232F9V9 E2AQ44 A0A3L8E234 A0A067R3T3 A0A291P102 A0A336MPQ2 A0A1B6EWY6 A0A336ME48 A0A1I8PT77 K7ITZ6 A0A2J7QLL4 A0A291P0R7 A0A158NWQ4 A0A2P8XMK6 A0A212F246 A0A1S3D8R6 Q178U1 A0A2S2QBR1 A0A437BLQ3 F4WQ30 A0A0A9ZH27 A0A151IHR1 A0A1A9VQK6 A0A023EV68 A0A194PXW0 A0A067QZE3 B4GMR4 D0QWK8 A0A1I8N555 A0A1A9Y074 A0A0R3NJP9 A0A1B0FAV7 A0A1A9Z6K6 A0A1Y1MP57 P18172 A0A026W0A3 A0A3B0JPT6 D6WN48 O44106 A0A151JUB7 A0A154P559 B4M4G1 A0A195EDG1 A0A1A9WL86 A0A1B0DAQ7 B3LYF6 Q8SXV0 V5GRM6 A0A151I5L3 B4I4R7 E9IVZ8 A0A0C9S068 U3PT72 Q94963 A0A2U8U0K3 P18173 B3P2V7 A0A0C9PXZ5 B4N874
B8XZY7 A0A194Q009 A0A2A4JD42 A0A218N0E8 A0A212FMI8 A0A2H1VAE8 A0A194RR25 A0A194Q0T0 A0A3S2LZ99 A0A2A4K284 A0A2S2NJV8 A0A182RS69 A0A2M4CT25 J9JWH7 D6WPB8 A0A182VBZ4 A0A182XAV1 A0A182LES2 Q7QCC5 A0A182HSP9 A0A182MWU5 A0A1Y1KHY8 A0A182VWW9 A0A182UL69 E0VK19 A0A139WGK8 A0A1Q3FML5 A0A182J274 A0A087ZQ05 B0WR76 A0A182NRL5 A0A2H1WCL9 A0A2A3EJY8 A0A182R0I2 A0A2J7Q741 A0A232F9V9 E2AQ44 A0A3L8E234 A0A067R3T3 A0A291P102 A0A336MPQ2 A0A1B6EWY6 A0A336ME48 A0A1I8PT77 K7ITZ6 A0A2J7QLL4 A0A291P0R7 A0A158NWQ4 A0A2P8XMK6 A0A212F246 A0A1S3D8R6 Q178U1 A0A2S2QBR1 A0A437BLQ3 F4WQ30 A0A0A9ZH27 A0A151IHR1 A0A1A9VQK6 A0A023EV68 A0A194PXW0 A0A067QZE3 B4GMR4 D0QWK8 A0A1I8N555 A0A1A9Y074 A0A0R3NJP9 A0A1B0FAV7 A0A1A9Z6K6 A0A1Y1MP57 P18172 A0A026W0A3 A0A3B0JPT6 D6WN48 O44106 A0A151JUB7 A0A154P559 B4M4G1 A0A195EDG1 A0A1A9WL86 A0A1B0DAQ7 B3LYF6 Q8SXV0 V5GRM6 A0A151I5L3 B4I4R7 E9IVZ8 A0A0C9S068 U3PT72 Q94963 A0A2U8U0K3 P18173 B3P2V7 A0A0C9PXZ5 B4N874
PDB
5NCC
E-value=1.52408e-63,
Score=618
Ontologies
PATHWAY
GO
Topology
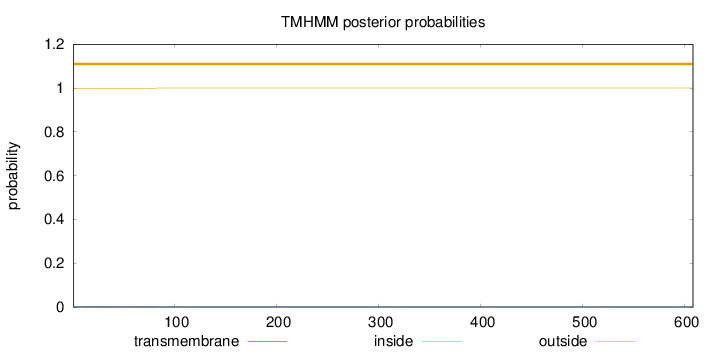
Subcellular location
Secreted
Length:
608
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0562000000000002
Exp number, first 60 AAs:
0.03272
Total prob of N-in:
0.00227
outside
1 - 608
Population Genetic Test Statistics
Pi
187.782129
Theta
170.163359
Tajima's D
1.016514
CLR
0.728715
CSRT
0.664766761661917
Interpretation
Uncertain
