Pre Gene Modal
BGIBMGA012963
Annotation
PREDICTED:_39S_ribosomal_protein_L44?_mitochondrial-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.992
Sequence
CDS
ATGGCACTAATGAGAAGATGTGCTCCGCTTTTTGCTAGGTGTATCAAAATACCACCTATGACTTTACAGAGTCAAAAAATTCATCGATGGGTAGCTCCTACGCTTGTAGAATTAAAGAGGAGAGAAAAAAAGTTAGGCGGAAAGAAAACAAATCCCCGTAACACTTTTCTGGAATGGAATTTGGAGGCTGAACTTTATGCTTTTGGAAAACGTTTGAATGAAGAATTCGACTCGGATTTGTTACTACAAGCGTTTACTGACAGAACTTATATTATAAAGGAAGAAATGAAACAAAAAGAAATGGGAGTAGATATTCCTATGAAGGACAATATGGAGTTGATTGCAGAAGGAGAAAAGTTCATAAATGAATATGTGCAGCTCTACCTAGAGACAGTTTTACCGAAATTTCCATTAGAGGGAGTGAATGAAGTGAGAAATTATCTTACAAATGAGGACAGACTGGCTAAAATCTCCTTGAGTCTGGGAACTAAGGACATCATACTAGCTGCAGAATATCCAGTTGATAATTACATTTTAGCAAACACTTTTAAAGCCATTGTAGCTGCTTTGTTAGAATCAACAAGTGAAGAAAGAGCAGCACATTTTGTGAGAGATTTTGTTATCACACAGTTACAAGGTAAAGATGTGAACGAGTATTGGAACATTGAAGACCCTTGGACTATGCTTACTGGCATCTTAGAAAAAGATGGAAGTAAAGTAGAACCTCGACTAATTGGAGAAGTTGGAAAAAATACGTTACTTGCCTGTTACAGAATTGGCCTTTATGTGAACAAGAAAATGATATCTGAAGGTTTCGGTGAAAAAATTGAAATAGCAAAAGAAATGGCTGCTAGAGAAGCTCTTAAAAGAATATTTGGTACCGAAGATCACATGAAACCTATTAATTTTAAAATTTCTGGTATACCAAAGGGGTCCTCCCAAGAACGTTATCAGATTGCCAGTAATTAA
Protein
MALMRRCAPLFARCIKIPPMTLQSQKIHRWVAPTLVELKRREKKLGGKKTNPRNTFLEWNLEAELYAFGKRLNEEFDSDLLLQAFTDRTYIIKEEMKQKEMGVDIPMKDNMELIAEGEKFINEYVQLYLETVLPKFPLEGVNEVRNYLTNEDRLAKISLSLGTKDIILAAEYPVDNYILANTFKAIVAALLESTSEERAAHFVRDFVITQLQGKDVNEYWNIEDPWTMLTGILEKDGSKVEPRLIGEVGKNTLLACYRIGLYVNKKMISEGFGEKIEIAKEMAAREALKRIFGTEDHMKPINFKISGIPKGSSQERYQIASN
Summary
Uniprot
A0A2H1VPZ4
A0A2A4JXT0
A0A1E1VZ71
S4PY42
A0A437B8K2
A0A212FME1
+ More
A0A194R2R9 A0A1E1WM95 A0A194PZZ6 A0A1B6DW47 A0A2J7PRF7 U5ES34 A0A067R8Z2 A0A1B0ETK1 C4WUR1 J9JM41 A0A2J7PRF0 A0A1Q3FDZ0 B0WMD0 A0A2H8TQA4 A0A182GX17 A0A2S2PIR6 Q16ZJ3 A0A336LLF6 A0A1B6JY34 A0A1L8DXP2 A0A2S2R1V5 A0A182XBL5 A0A182UUU1 A0A1B6G8Q0 A0A1B0CP42 Q7PYE3 A0A1B6ESP8 A0A1L8DXM3 A0A182I0F1 A0A182QZU8 A0A182VQY1 A0A182YJT9 A0A1J1IM11 A0A1B6MAC5 A0A182UH00 A0A182LUW0 A0A182JQR9 A0A182N5X8 W5J4N7 A0A182PC26 A0A0L0BUM4 A0A182IW67 A0A1I8PDM5 A0A1B0DLK7 A0A0A1WS74 A0A182R1J0 A0A1I8N5K1 A0A182FQK2 A0A2M4APV5 A0A0K8TSF3 A0A2M4AQ62 A0A1A9VPK7 A0A0V0GBN8 A0A069DRK0 D3TMI3 E0VMZ1 A0A0K8UE72 A0A1W4UJH9 A0A1W4UY64 T1I3F3 A0A151ID31 W8C4R2 A0A1A9WDE8 A0A0M4F4I4 B4LZC2 A0A158N9M6 A0A0J7NHB2 A0A034WTV2 A0A0P4VLJ3 A0A0Q9WGN1 B4NB90 E2C0W5 A0A151ISQ9 A0A1A9XU25 A0A146LTI7 A0A0K8TIJ7 B3LZH8 A0A0A9YSG8 B4JHV7 A0A195F8G3 A0A3B0JQF6 Q9VNC1 Q7K141 A0A2P8XUL6 A0A232F227 B4I407 B4QWW0 F4WTR6 A0A1D2MY72 A0A3R7NDS2 A0A195BNK3 A0A026W3R7 B4KD97
A0A194R2R9 A0A1E1WM95 A0A194PZZ6 A0A1B6DW47 A0A2J7PRF7 U5ES34 A0A067R8Z2 A0A1B0ETK1 C4WUR1 J9JM41 A0A2J7PRF0 A0A1Q3FDZ0 B0WMD0 A0A2H8TQA4 A0A182GX17 A0A2S2PIR6 Q16ZJ3 A0A336LLF6 A0A1B6JY34 A0A1L8DXP2 A0A2S2R1V5 A0A182XBL5 A0A182UUU1 A0A1B6G8Q0 A0A1B0CP42 Q7PYE3 A0A1B6ESP8 A0A1L8DXM3 A0A182I0F1 A0A182QZU8 A0A182VQY1 A0A182YJT9 A0A1J1IM11 A0A1B6MAC5 A0A182UH00 A0A182LUW0 A0A182JQR9 A0A182N5X8 W5J4N7 A0A182PC26 A0A0L0BUM4 A0A182IW67 A0A1I8PDM5 A0A1B0DLK7 A0A0A1WS74 A0A182R1J0 A0A1I8N5K1 A0A182FQK2 A0A2M4APV5 A0A0K8TSF3 A0A2M4AQ62 A0A1A9VPK7 A0A0V0GBN8 A0A069DRK0 D3TMI3 E0VMZ1 A0A0K8UE72 A0A1W4UJH9 A0A1W4UY64 T1I3F3 A0A151ID31 W8C4R2 A0A1A9WDE8 A0A0M4F4I4 B4LZC2 A0A158N9M6 A0A0J7NHB2 A0A034WTV2 A0A0P4VLJ3 A0A0Q9WGN1 B4NB90 E2C0W5 A0A151ISQ9 A0A1A9XU25 A0A146LTI7 A0A0K8TIJ7 B3LZH8 A0A0A9YSG8 B4JHV7 A0A195F8G3 A0A3B0JQF6 Q9VNC1 Q7K141 A0A2P8XUL6 A0A232F227 B4I407 B4QWW0 F4WTR6 A0A1D2MY72 A0A3R7NDS2 A0A195BNK3 A0A026W3R7 B4KD97
Pubmed
23622113
22118469
26354079
24845553
26483478
17510324
+ More
12364791 14747013 17210077 25244985 20920257 23761445 26108605 25830018 25315136 26369729 26334808 20353571 20566863 24495485 17994087 21347285 25348373 27129103 20798317 26823975 25401762 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 29403074 28648823 21719571 27289101 24508170 30249741
12364791 14747013 17210077 25244985 20920257 23761445 26108605 25830018 25315136 26369729 26334808 20353571 20566863 24495485 17994087 21347285 25348373 27129103 20798317 26823975 25401762 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 29403074 28648823 21719571 27289101 24508170 30249741
EMBL
ODYU01003753
SOQ42915.1
NWSH01000463
PCG76230.1
GDQN01011103
JAT79951.1
+ More
GAIX01003943 JAA88617.1 RSAL01000115 RVE46876.1 AGBW02007651 OWR54934.1 KQ460878 KPJ11545.1 GDQN01003073 JAT87981.1 KQ459582 KPI98902.1 GEDC01007439 JAS29859.1 NEVH01022362 PNF18914.1 GANO01003428 JAB56443.1 KK852622 KDR19986.1 AJWK01010554 AK341177 BAH71631.1 ABLF02036417 PNF18913.1 GFDL01009333 JAV25712.1 DS231997 EDS30987.1 GFXV01003633 MBW15438.1 JXUM01094434 KQ564128 KXJ72728.1 GGMR01016704 MBY29323.1 CH477489 EAT40081.1 UFQT01000043 SSX18786.1 GECU01003598 JAT04109.1 GFDF01002873 JAV11211.1 GGMS01014763 MBY83966.1 GECZ01010944 JAS58825.1 AJWK01021412 AAAB01008987 EAA01591.4 GECZ01029036 JAS40733.1 GFDF01002874 JAV11210.1 APCN01002072 AXCN02000191 CVRI01000051 CRK99505.1 GEBQ01007173 JAT32804.1 AXCM01001692 ADMH02002134 ETN58383.1 JRES01001304 KNC23742.1 AJVK01036466 GBXI01012962 JAD01330.1 GGFK01009493 MBW42814.1 GDAI01000537 JAI17066.1 GGFK01009600 MBW42921.1 GECL01000620 JAP05504.1 GBGD01002344 JAC86545.1 CCAG010002244 EZ422635 ADD18911.1 DS235331 EEB14747.1 GDHF01027664 JAI24650.1 ACPB03023076 KQ977991 KYM98302.1 GAMC01005334 GAMC01005330 JAC01222.1 CP012526 ALC46650.1 CH940650 EDW68157.1 ADTU01009704 LBMM01005014 KMQ91900.1 GAKP01000873 JAC58079.1 GDKW01003063 JAI53532.1 KRF83709.1 CH964232 EDW81054.1 GL451850 EFN78401.1 KQ981064 KYN09902.1 GDHC01008547 GDHC01004895 JAQ10082.1 JAQ13734.1 GBRD01000861 JAG64960.1 CH902617 EDV41920.1 GBHO01009038 JAG34566.1 CH916369 EDV92865.1 KQ981727 KYN36671.1 OUUW01000008 SPP84365.1 AE014297 AAF52025.1 AY069395 AAL39540.2 PYGN01001321 PSN35695.1 NNAY01001280 OXU24508.1 CH480821 EDW54950.1 CM000364 EDX11722.1 GL888344 EGI62381.1 LJIJ01000394 ODM97938.1 QCYY01000537 ROT84453.1 KQ976428 KYM87802.1 KK107502 QOIP01000014 EZA49689.1 RLU14875.1 CH933806 EDW14879.1
GAIX01003943 JAA88617.1 RSAL01000115 RVE46876.1 AGBW02007651 OWR54934.1 KQ460878 KPJ11545.1 GDQN01003073 JAT87981.1 KQ459582 KPI98902.1 GEDC01007439 JAS29859.1 NEVH01022362 PNF18914.1 GANO01003428 JAB56443.1 KK852622 KDR19986.1 AJWK01010554 AK341177 BAH71631.1 ABLF02036417 PNF18913.1 GFDL01009333 JAV25712.1 DS231997 EDS30987.1 GFXV01003633 MBW15438.1 JXUM01094434 KQ564128 KXJ72728.1 GGMR01016704 MBY29323.1 CH477489 EAT40081.1 UFQT01000043 SSX18786.1 GECU01003598 JAT04109.1 GFDF01002873 JAV11211.1 GGMS01014763 MBY83966.1 GECZ01010944 JAS58825.1 AJWK01021412 AAAB01008987 EAA01591.4 GECZ01029036 JAS40733.1 GFDF01002874 JAV11210.1 APCN01002072 AXCN02000191 CVRI01000051 CRK99505.1 GEBQ01007173 JAT32804.1 AXCM01001692 ADMH02002134 ETN58383.1 JRES01001304 KNC23742.1 AJVK01036466 GBXI01012962 JAD01330.1 GGFK01009493 MBW42814.1 GDAI01000537 JAI17066.1 GGFK01009600 MBW42921.1 GECL01000620 JAP05504.1 GBGD01002344 JAC86545.1 CCAG010002244 EZ422635 ADD18911.1 DS235331 EEB14747.1 GDHF01027664 JAI24650.1 ACPB03023076 KQ977991 KYM98302.1 GAMC01005334 GAMC01005330 JAC01222.1 CP012526 ALC46650.1 CH940650 EDW68157.1 ADTU01009704 LBMM01005014 KMQ91900.1 GAKP01000873 JAC58079.1 GDKW01003063 JAI53532.1 KRF83709.1 CH964232 EDW81054.1 GL451850 EFN78401.1 KQ981064 KYN09902.1 GDHC01008547 GDHC01004895 JAQ10082.1 JAQ13734.1 GBRD01000861 JAG64960.1 CH902617 EDV41920.1 GBHO01009038 JAG34566.1 CH916369 EDV92865.1 KQ981727 KYN36671.1 OUUW01000008 SPP84365.1 AE014297 AAF52025.1 AY069395 AAL39540.2 PYGN01001321 PSN35695.1 NNAY01001280 OXU24508.1 CH480821 EDW54950.1 CM000364 EDX11722.1 GL888344 EGI62381.1 LJIJ01000394 ODM97938.1 QCYY01000537 ROT84453.1 KQ976428 KYM87802.1 KK107502 QOIP01000014 EZA49689.1 RLU14875.1 CH933806 EDW14879.1
Proteomes
UP000218220
UP000283053
UP000007151
UP000053240
UP000053268
UP000235965
+ More
UP000027135 UP000092461 UP000007819 UP000002320 UP000069940 UP000249989 UP000008820 UP000076407 UP000075903 UP000007062 UP000075840 UP000075886 UP000075920 UP000076408 UP000183832 UP000075902 UP000075883 UP000075881 UP000075884 UP000000673 UP000075885 UP000037069 UP000075880 UP000095300 UP000092462 UP000075900 UP000095301 UP000069272 UP000078200 UP000092444 UP000009046 UP000192221 UP000015103 UP000078542 UP000091820 UP000092553 UP000008792 UP000005205 UP000036403 UP000007798 UP000008237 UP000078492 UP000092443 UP000007801 UP000001070 UP000078541 UP000268350 UP000000803 UP000245037 UP000215335 UP000001292 UP000000304 UP000007755 UP000094527 UP000283509 UP000078540 UP000053097 UP000279307 UP000009192
UP000027135 UP000092461 UP000007819 UP000002320 UP000069940 UP000249989 UP000008820 UP000076407 UP000075903 UP000007062 UP000075840 UP000075886 UP000075920 UP000076408 UP000183832 UP000075902 UP000075883 UP000075881 UP000075884 UP000000673 UP000075885 UP000037069 UP000075880 UP000095300 UP000092462 UP000075900 UP000095301 UP000069272 UP000078200 UP000092444 UP000009046 UP000192221 UP000015103 UP000078542 UP000091820 UP000092553 UP000008792 UP000005205 UP000036403 UP000007798 UP000008237 UP000078492 UP000092443 UP000007801 UP000001070 UP000078541 UP000268350 UP000000803 UP000245037 UP000215335 UP000001292 UP000000304 UP000007755 UP000094527 UP000283509 UP000078540 UP000053097 UP000279307 UP000009192
Pfam
Interpro
CDD
ProteinModelPortal
A0A2H1VPZ4
A0A2A4JXT0
A0A1E1VZ71
S4PY42
A0A437B8K2
A0A212FME1
+ More
A0A194R2R9 A0A1E1WM95 A0A194PZZ6 A0A1B6DW47 A0A2J7PRF7 U5ES34 A0A067R8Z2 A0A1B0ETK1 C4WUR1 J9JM41 A0A2J7PRF0 A0A1Q3FDZ0 B0WMD0 A0A2H8TQA4 A0A182GX17 A0A2S2PIR6 Q16ZJ3 A0A336LLF6 A0A1B6JY34 A0A1L8DXP2 A0A2S2R1V5 A0A182XBL5 A0A182UUU1 A0A1B6G8Q0 A0A1B0CP42 Q7PYE3 A0A1B6ESP8 A0A1L8DXM3 A0A182I0F1 A0A182QZU8 A0A182VQY1 A0A182YJT9 A0A1J1IM11 A0A1B6MAC5 A0A182UH00 A0A182LUW0 A0A182JQR9 A0A182N5X8 W5J4N7 A0A182PC26 A0A0L0BUM4 A0A182IW67 A0A1I8PDM5 A0A1B0DLK7 A0A0A1WS74 A0A182R1J0 A0A1I8N5K1 A0A182FQK2 A0A2M4APV5 A0A0K8TSF3 A0A2M4AQ62 A0A1A9VPK7 A0A0V0GBN8 A0A069DRK0 D3TMI3 E0VMZ1 A0A0K8UE72 A0A1W4UJH9 A0A1W4UY64 T1I3F3 A0A151ID31 W8C4R2 A0A1A9WDE8 A0A0M4F4I4 B4LZC2 A0A158N9M6 A0A0J7NHB2 A0A034WTV2 A0A0P4VLJ3 A0A0Q9WGN1 B4NB90 E2C0W5 A0A151ISQ9 A0A1A9XU25 A0A146LTI7 A0A0K8TIJ7 B3LZH8 A0A0A9YSG8 B4JHV7 A0A195F8G3 A0A3B0JQF6 Q9VNC1 Q7K141 A0A2P8XUL6 A0A232F227 B4I407 B4QWW0 F4WTR6 A0A1D2MY72 A0A3R7NDS2 A0A195BNK3 A0A026W3R7 B4KD97
A0A194R2R9 A0A1E1WM95 A0A194PZZ6 A0A1B6DW47 A0A2J7PRF7 U5ES34 A0A067R8Z2 A0A1B0ETK1 C4WUR1 J9JM41 A0A2J7PRF0 A0A1Q3FDZ0 B0WMD0 A0A2H8TQA4 A0A182GX17 A0A2S2PIR6 Q16ZJ3 A0A336LLF6 A0A1B6JY34 A0A1L8DXP2 A0A2S2R1V5 A0A182XBL5 A0A182UUU1 A0A1B6G8Q0 A0A1B0CP42 Q7PYE3 A0A1B6ESP8 A0A1L8DXM3 A0A182I0F1 A0A182QZU8 A0A182VQY1 A0A182YJT9 A0A1J1IM11 A0A1B6MAC5 A0A182UH00 A0A182LUW0 A0A182JQR9 A0A182N5X8 W5J4N7 A0A182PC26 A0A0L0BUM4 A0A182IW67 A0A1I8PDM5 A0A1B0DLK7 A0A0A1WS74 A0A182R1J0 A0A1I8N5K1 A0A182FQK2 A0A2M4APV5 A0A0K8TSF3 A0A2M4AQ62 A0A1A9VPK7 A0A0V0GBN8 A0A069DRK0 D3TMI3 E0VMZ1 A0A0K8UE72 A0A1W4UJH9 A0A1W4UY64 T1I3F3 A0A151ID31 W8C4R2 A0A1A9WDE8 A0A0M4F4I4 B4LZC2 A0A158N9M6 A0A0J7NHB2 A0A034WTV2 A0A0P4VLJ3 A0A0Q9WGN1 B4NB90 E2C0W5 A0A151ISQ9 A0A1A9XU25 A0A146LTI7 A0A0K8TIJ7 B3LZH8 A0A0A9YSG8 B4JHV7 A0A195F8G3 A0A3B0JQF6 Q9VNC1 Q7K141 A0A2P8XUL6 A0A232F227 B4I407 B4QWW0 F4WTR6 A0A1D2MY72 A0A3R7NDS2 A0A195BNK3 A0A026W3R7 B4KD97
PDB
6GB2
E-value=8.60276e-47,
Score=470
Ontologies
GO
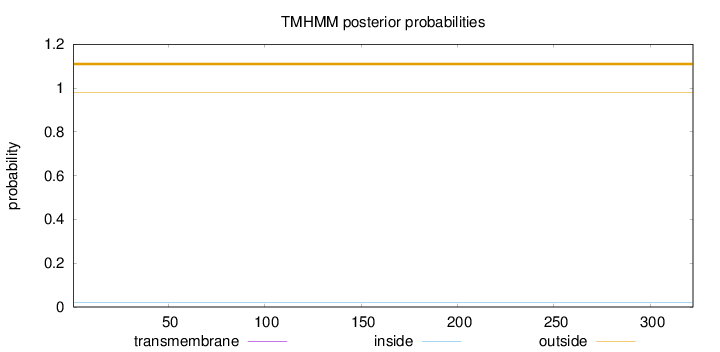
Topology
Length:
322
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00277
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.01946
outside
1 - 322
Population Genetic Test Statistics
Pi
270.366657
Theta
206.487047
Tajima's D
0.998088
CLR
0.24109
CSRT
0.662816859157042
Interpretation
Uncertain
